An Improved F(st) Estimator
- PMID: 26317214
- PMCID: PMC4552798
- DOI: 10.1371/journal.pone.0135368
An Improved F(st) Estimator
Abstract
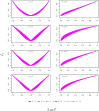
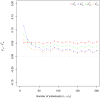
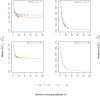
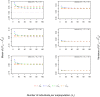
The fixation index F(st) plays a central role in ecological and evolutionary genetic studies. The estimators of Wright ([Formula: see text]), Weir and Cockerham ([Formula: see text]), and Hudson et al. ([Formula: see text]) are widely used to measure genetic differences among different populations, but all have limitations. We propose a minimum variance estimator [Formula: see text] using [Formula: see text] and [Formula: see text]. We tested [Formula: see text] in simulations and applied it to 120 unrelated East African individuals from Ethiopia and 11 subpopulations in HapMap 3 with 464,642 SNPs. Our simulation study showed that [Formula: see text] has smaller bias than [Formula: see text] for small sample sizes and smaller bias than [Formula: see text] for large sample sizes. Also, [Formula: see text] has smaller variance than [Formula: see text] for small Fst values and smaller variance than [Formula: see text] for large F(st) values. We demonstrated that approximately 30 subpopulations and 30 individuals per subpopulation are required in order to accurately estimate F(st).
Conflict of interest statement
Figures
References
-
- Wright S. The genetical structure of populations. Ann Eugen 1951; 15(4): 323–354. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

