Comparative analysis of tools for live cell imaging of actin network architecture
- PMID: 26317264
- PMCID: PMC4914014
- DOI: 10.1080/19490992.2014.1047714
Comparative analysis of tools for live cell imaging of actin network architecture
Abstract
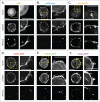
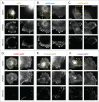
Fluorescent derivatives of actin and actin-binding domains are powerful tools for studying actin filament architecture and dynamics in live cells. Growing evidence, however, indicates that these probes are biased, and their cellular distribution does not accurately reflect that of the cytoskeleton. To understand the strengths and weaknesses of commonly used live-cell probes--fluorescent protein fusions of actin, Lifeact, F-tractin, and actin-binding domains from utrophin--we compared their distributions in cells derived from various model organisms. We focused on five actin networks: the peripheral cortex, lamellipodial and lamellar networks, filopodial bundles, and stress fibers. Using phalloidin as a standard, we identified consistent biases in the distribution of each probe. The localization of F-tractin is the most similar to that of phalloidin but induces organism-specific changes in cell morphology. Both Lifeact and GFP-actin concentrate in lamellipodial actin networks but are excluded from lamellar networks and filopodia. In contrast, the full utrophin actin-binding domain (Utr261) binds filaments of the lamellum but only weakly localizes to lamellipodia, while a shorter variant (Utr230) is restricted to the most stable subpopulations of actin filaments: cortical networks and stress fibers. In some cells, Utr230 also detects Golgi-associated filaments, previously detected by immunofluorescence but not visible by phalloidin staining. Consistent with its localization, Utr230 exhibits slow rates of fluorescence recovery after photobleaching (FRAP) compared to F-tractin, Utr261 and Lifeact, suggesting that it may be more useful for FRAP- and photo-activation-based studies of actin network dynamics.
Keywords: Cytoskeleton; actin; cell architecture; fluorescent protein reporters; live cell imaging.
Figures






References
-
- De La Cruz EM, Pollard TD. Transient kinetic analysis of rhodamine phalloidin binding to actin filaments. Biochemistry 1994; 33:14387-14392 - PubMed
-
- Schell MJ, Erneux C., Irvine RF. Inositol 1,4,5-trisphosphate 3-kinase A associates with F-actin and dendritic spines via its N terminus. J. Biol. Chem. 2001; 276(40):37537-46 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
