miR-155, identified as anti-metastatic by global miRNA profiling of a metastasis model, inhibits cancer cell extravasation and colonization in vivo and causes significant signaling alterations
- PMID: 26317550
- PMCID: PMC4745722
- DOI: 10.18632/oncotarget.4942
miR-155, identified as anti-metastatic by global miRNA profiling of a metastasis model, inhibits cancer cell extravasation and colonization in vivo and causes significant signaling alterations
Abstract
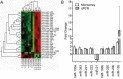
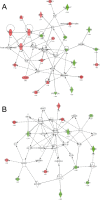
To gain insight into miRNA regulation in metastasis formation, we used a metastasis cell line model that allows investigation of extravasation and colonization of circulating cancer cells to lungs in mice. Using global miRNA profiling, 28 miRNAs were found to exhibit significantly altered expression between isogenic metastasizing and non-metastasizing cancer cells, with miR-155 being the most differentially expressed. Highly metastatic mesenchymal-like CL16 cancer cells showed very low miR-155 expression, and miR-155 overexpression in these cells lead to significantly decreased tumor burden in lungs when injected intravenously in immunodeficient mice. Our experiments addressing the underlying mechanism of the altered tumor burden revealed that miR-155-overexpressing CL16 cells were less invasive than CL16 control cells in vitro, while miR-155 overexpression had no effect on cancer cell proliferation or apoptosis in established lung tumors. To identify proteins regulated by miR-155 and thus delineate its function in our cell model, we compared the proteome of xenograft tumors derived from miR-155-overexpressing CL16 cells and CL16 control cells using mass spectrometry-based proteomics. >4,000 proteins were identified, of which 92 were consistently differentially expressed. Network analysis revealed that the altered proteins were associated with cellular functions such as movement, growth and survival as well as cell-to-cell signaling and interaction. Downregulation of the three metastasis-associated proteins ALDH1A1, PIR and PDCD4 in miR-155-overexpressing tumors was validated by immunohistochemistry. Our results demonstrate that miR-155 inhibits the ability of cancer cells to extravasate and/or colonize at distant organs and brings additional insight into the complexity of miR-155 regulation in metastatic seeding.
Keywords: LNA miRNA microarray; cancer; colonization; in vivo metastasis cell line model; miR-155.
Conflict of interest statement
The authors declare no conflicts of interests
Figures




Similar articles
-
NADH-Cytochrome b5 Reductase 3 Promotes Colonization and Metastasis Formation and Is a Prognostic Marker of Disease-Free and Overall Survival in Estrogen Receptor-Negative Breast Cancer.Mol Cell Proteomics. 2015 Nov;14(11):2988-99. doi: 10.1074/mcp.M115.050385. Epub 2015 Sep 8. Mol Cell Proteomics. 2015. PMID: 26351264 Free PMC article.
-
MicroRNA 744-3p promotes MMP-9-mediated metastasis by simultaneously suppressing PDCD4 and PTEN in laryngeal squamous cell carcinoma.Oncotarget. 2016 Sep 6;7(36):58218-58233. doi: 10.18632/oncotarget.11280. Oncotarget. 2016. PMID: 27533461 Free PMC article.
-
Hypoxic BMSC-derived exosomal miRNAs promote metastasis of lung cancer cells via STAT3-induced EMT.Mol Cancer. 2019 Mar 13;18(1):40. doi: 10.1186/s12943-019-0959-5. Mol Cancer. 2019. PMID: 30866952 Free PMC article.
-
Tumor-Progressive Mechanisms Mediating miRNA-Protein Interaction.Int J Mol Sci. 2021 Nov 14;22(22):12303. doi: 10.3390/ijms222212303. Int J Mol Sci. 2021. PMID: 34830186 Free PMC article. Review.
-
Proteome and miRNome profiling of microvesicles derived from medulloblastoma cell lines with stem-like properties reveals biomarkers of poor prognosis.Brain Res. 2020 Mar 1;1730:146646. doi: 10.1016/j.brainres.2020.146646. Epub 2020 Jan 7. Brain Res. 2020. PMID: 31917138 Review.
Cited by
-
Targeting microRNAs: a new action mechanism of natural compounds.Oncotarget. 2017 Feb 28;8(9):15961-15970. doi: 10.18632/oncotarget.14392. Oncotarget. 2017. PMID: 28052018 Free PMC article. Review.
-
Regulators at Every Step-How microRNAs Drive Tumor Cell Invasiveness and Metastasis.Cancers (Basel). 2020 Dec 10;12(12):3709. doi: 10.3390/cancers12123709. Cancers (Basel). 2020. PMID: 33321819 Free PMC article. Review.
-
A Feedback Loop between MicroRNA 155 (miR-155), Programmed Cell Death 4, and Activation Protein 1 Modulates the Expression of miR-155 and Tumorigenesis in Tongue Cancer.Mol Cell Biol. 2019 Mar 1;39(6):e00410-18. doi: 10.1128/MCB.00410-18. Print 2019 Mar 15. Mol Cell Biol. 2019. PMID: 30617160 Free PMC article.
-
Inflammation as a risk factor for stroke in atrial fibrillation: data from a microarray data analysis.J Int Med Res. 2020 May;48(5):300060520921671. doi: 10.1177/0300060520921671. J Int Med Res. 2020. PMID: 32367757 Free PMC article.
-
A new era of cancer immunotherapy: vaccines and miRNAs.Cancer Immunol Immunother. 2025 Apr 1;74(5):163. doi: 10.1007/s00262-025-04011-5. Cancer Immunol Immunother. 2025. PMID: 40167762 Free PMC article. Review.
References
-
- Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2:563–572. - PubMed
-
- Weigelt B, Peterse JL, van 't Veer LJ. Breast cancer metastasis: markers and models. Nat Rev Cancer. 2005;5:591–602. - PubMed
-
- White NM, Fatoohi E, Metias M, Jung K, Stephan C, Yousef GM. Metastamirs: a stepping stone towards improved cancer management. Nat Rev Clin Oncol. 2011;8:75–84. - PubMed
-
- Tsuchiya S, Okuno Y, Tsujimoto G. MicroRNA: biogenetic and functional mechanisms and involvements in cell differentiation and cancer. J Pharmacol Sci. 2006;101:267–270. - PubMed
-
- Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Ménard S, Palazzo JP, Rosenberg A, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

