Automated tumor analysis for molecular profiling in lung cancer
- PMID: 26317646
- PMCID: PMC4695036
- DOI: 10.18632/oncotarget.4391
Automated tumor analysis for molecular profiling in lung cancer
Abstract
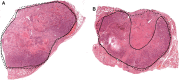
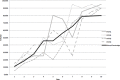
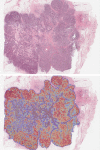
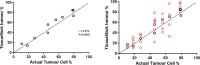
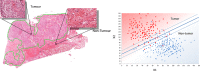
The discovery and clinical application of molecular biomarkers in solid tumors, increasingly relies on nucleic acid extraction from FFPE tissue sections and subsequent molecular profiling. This in turn requires the pathological review of haematoxylin & eosin (H&E) stained slides, to ensure sample quality, tumor DNA sufficiency by visually estimating the percentage tumor nuclei and tumor annotation for manual macrodissection. In this study on NSCLC, we demonstrate considerable variation in tumor nuclei percentage between pathologists, potentially undermining the precision of NSCLC molecular evaluation and emphasising the need for quantitative tumor evaluation. We subsequently describe the development and validation of a system called TissueMark for automated tumor annotation and percentage tumor nuclei measurement in NSCLC using computerized image analysis. Evaluation of 245 NSCLC slides showed precise automated tumor annotation of cases using Tissuemark, strong concordance with manually drawn boundaries and identical EGFR mutational status, following manual macrodissection from the image analysis generated tumor boundaries. Automated analysis of cell counts for % tumor measurements by Tissuemark showed reduced variability and significant correlation (p < 0.001) with benchmark tumor cell counts. This study demonstrates a robust image analysis technology that can facilitate the automated quantitative analysis of tissue samples for molecular profiling in discovery and diagnostics.
Keywords: digital pathology; image analysis; manual macrodissection; molecular pathology; percentage tumor.
Conflict of interest statement
Professor Peter Hamilton is the founder of and non-executive director with PathXL Ltd. Jonathon Tunstall is the Director of Product Strategy with PathXL Ltd. David McCleary is Head of Development and Research at PathXL. Jim Diamond is Research Lead at PathXL Ltd. Professor Manuel Salto-Tellez is a Senior Advisor to PathXL.
Figures





References
-
- Amir-Aslani A, Mangematin V. The future of drug discovery and development: Shifting emphasis towards personalized medicine. Technol. Forecast. Soc. Change. 2010;77:203–17.
-
- Kasaian K, Jones SJ. A new frontier in personalized cancer therapy: mapping molecular changes. Future Oncol. 2011;7:873–94. - PubMed
-
- Mirnezami R, Nicholson J, Darzi A. Preparing for Precision Medicine. N. Engl. J. Med. 2012:489–91. - PubMed
-
- Smits AJJ, Kummer JA, de Bruin PC, Bol M, van den Tweel JG, Seldenrijk K a, et al. The estimation of tumor cell percentage for molecular testing by pathologists is not accurate. Mod. Pathol. 2014;27:168–74. [Internet]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23887293. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

