Development of a scalable suspension culture for cardiac differentiation from human pluripotent stem cells
- PMID: 26318718
- PMCID: PMC4600677
- DOI: 10.1016/j.scr.2015.08.002
Development of a scalable suspension culture for cardiac differentiation from human pluripotent stem cells
Abstract
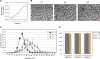
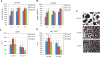
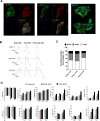
To meet the need of a large quantity of hPSC-derived cardiomyocytes (CM) for pre-clinical and clinical studies, a robust and scalable differentiation system for CM production is essential. With a human pluripotent stem cells (hPSC) aggregate suspension culture system we established previously, we developed a matrix-free, scalable, and GMP-compliant process for directing hPSC differentiation to CM in suspension culture by modulating Wnt pathways with small molecules. By optimizing critical process parameters including: cell aggregate size, small molecule concentrations, induction timing, and agitation rate, we were able to consistently differentiate hPSCs to >90% CM purity with an average yield of 1.5 to 2×10(9) CM/L at scales up to 1L spinner flasks. CM generated from the suspension culture displayed typical genetic, morphological, and electrophysiological cardiac cell characteristics. This suspension culture system allows seamless transition from hPSC expansion to CM differentiation in a continuous suspension culture. It not only provides a cost and labor effective scalable process for large scale CM production, but also provides a bioreactor prototype for automation of cell manufacturing, which will accelerate the advance of hPSC research towards therapeutic applications.
Keywords: Cardiomyocyte differentiation; GMP; Human pluripotent stem cells; Suspension cell cultures.
Copyright © 2015. Published by Elsevier B.V.
Figures




References
-
- Arnold SJ, Robertson EJ. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nature reviews Molecular cell biology. 2009;10:91–103. - PubMed
-
- Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nature reviews Genetics. 2005;6:826–835. - PubMed
-
- Tam PP, Loebel DA. Gene function in mouse embryogenesis: get set for gastrulation. Nature reviews Genetics. 2007;8:368–381. - PubMed
-
- David R, et al. MesP1 drives vertebrate cardiovascular differentiation through Dkk-1-mediated blockade of Wnt-signalling. Nature cell biology. 2008;10:338–345. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

