The evolution of the dystroglycan complex, a major mediator of muscle integrity
- PMID: 26319583
- PMCID: PMC4582122
- DOI: 10.1242/bio.012468
The evolution of the dystroglycan complex, a major mediator of muscle integrity
Abstract
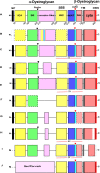
Basement membrane (BM) extracellular matrices are crucial for the coordination of different tissue layers. A matrix adhesion receptor that is important for BM function and stability in many mammalian tissues is the dystroglycan (DG) complex. This comprises the non-covalently-associated extracellular α-DG, that interacts with laminin in the BM, and the transmembrane β-DG, that interacts principally with dystrophin to connect to the actin cytoskeleton. Mutations in dystrophin, DG, or several enzymes that glycosylate α-DG underlie severe forms of human muscular dystrophy. Nonwithstanding the pathophysiological importance of the DG complex and its fundamental interest as a non-integrin system of cell-ECM adhesion, the evolution of DG and its interacting proteins is not understood. We analysed the phylogenetic distribution of DG, its proximal binding partners and key processing enzymes in extant metazoan and relevant outgroups. We identify that DG originated after the divergence of ctenophores from porifera and eumetazoa. The C-terminal half of the DG core protein is highly-conserved, yet the N-terminal region, that includes the laminin-binding region, has undergone major lineage-specific divergences. Phylogenetic analysis based on the C-terminal IG2_MAT_NU region identified three distinct clades corresponding to deuterostomes, arthropods, and mollusks/early-diverging metazoans. Whereas the glycosyltransferases that modify α-DG are also present in choanoflagellates, the DG-binding proteins dystrophin and laminin originated at the base of the metazoa, and DG-associated sarcoglycan is restricted to cnidarians and bilaterians. These findings implicate extensive functional diversification of DG within invertebrate lineages and identify the laminin-DG-dystrophin axis as a conserved adhesion system that evolved subsequent to integrin-ECM adhesion, likely to enhance the functional complexity of cell-BM interactions in early metazoans.
Keywords: Basement membrane; Dystroglycan; Dystroglycanopathy; Metazoa; Multicellularity; Protein domain analysis.
© 2015. Published by The Company of Biologists Ltd.
Conflict of interest statement
The authors declare no competing or financial interests.
Figures






References
-
- Adams J. C. (2013). The evolution of extracellular matrix: an over-view. In Evolution of Extracellular Matrix (ed. Keeley F. W. and Mecham R. P.), pp. 1-25. Berlin: Springer; 10.1007/978-3-642-36002-2 - DOI
-
- Bogdanik L., Framery B., Frölich A., Franco B., Mornet D., Bockaert J., Sigrist S. J., Grau Y. and Parmentier M.-L. (2008). Muscle dystroglycan organizes the postsynapse and regulates presynaptic neurotransmitter release at the Drosophila neuromuscular junction. PLoS ONE 3, e2084 10.1371/journal.pone.0002084 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

