Suppression of the GTPase-activating protein RGS10 increases Rheb-GTP and mTOR signaling in ovarian cancer cells
- PMID: 26319900
- PMCID: PMC4654121
- DOI: 10.1016/j.canlet.2015.08.012
Suppression of the GTPase-activating protein RGS10 increases Rheb-GTP and mTOR signaling in ovarian cancer cells
Abstract
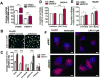
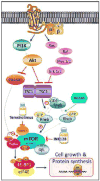
The regulator of G protein signaling 10 (RGS10) protein is a GTPase activating protein that accelerates the hydrolysis of GTP and therefore canonically inactivates G proteins, ultimately terminating signaling. Rheb is a small GTPase protein that shuttles between its GDP- and GTP-bound forms to activate mTOR. Since RGS10 suppression augments ovarian cancer cell viability, we sought to elucidate the molecular mechanism. Following RGS10 suppression in serum-free conditions, phosphorylation of mTOR, the eukaryotic translation initiation factor 4E binding protein 1 (4E-BP1), p70S6K and S6 Ribosomal Protein appear. Furthermore, suppressing RGS10 increases activated Rheb, suggesting RGS10 antagonizes mTOR signaling via the small G-protein. The effects of RGS10 suppression are enhanced after stimulating cells with the growth factor, lysophosphatidic acid, and reduced with mTOR inhibitors, temsirolimus and INK-128. Suppression of RGS10 leads to an increase in cell proliferation, even in the presence of etoposide. In summary, the RGS10 suppression increases Rheb-GTP and mTOR signaling in ovarian cancer cells. Our results suggest that RGS10 could serve in a novel, and previously unknown, role by accelerating the hydrolysis of GTP from Rheb in ovarian cancer cells.
Keywords: 4E-BP1; Lysophosphatidic acid; Regulator of G protein Signaling 10 protein (RGS10); Rheb; mTOR.
Copyright © 2015 Elsevier Ireland Ltd. All rights reserved.
Conflict of interest statement
The authors declare no conflicts of interest
Figures


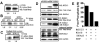

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

