HANABA TARANU regulates the shoot apical meristem and leaf development in cucumber (Cucumis sativus L.)
- PMID: 26320238
- PMCID: PMC4765787
- DOI: 10.1093/jxb/erv409
HANABA TARANU regulates the shoot apical meristem and leaf development in cucumber (Cucumis sativus L.)
Abstract
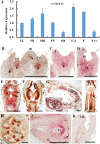
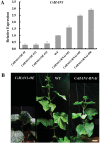
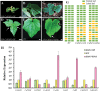
The shoot apical meristem (SAM) is essential for continuous organogenesis in higher plants, while the leaf is the primary source organ and the leaf shape directly affects the efficiency of photosynthesis. HANABA TARANU (HAN) encodes a GATA3-type transcription factor that functions in floral organ development, SAM organization, and embryo development in Arabidopsis, but is involved in suppressing bract outgrowth and promoting branching in grass species. Here the function of the HAN homologue CsHAN1 was characterized in cucumber, an important vegetable with great agricultural and economic value. CsHAN1 is predominantly expressed at the junction of the SAM and the stem, and can partially rescue the han-2 floral organ phenotype in Arabidopsis. Overexpression and RNAi of CsHAN1 transgenic cucumber resulted in retarded growth early after embryogenesis and produced highly lobed leaves. Further, it was found that CsHAN1 may regulate SAM development through regulating the WUSCHEL (WUS) and SHOOT MERISTEMLESS (STM) pathways, and mediate leaf development through a complicated gene regulatory network in cucumber.
Keywords: CsHAN; CsSTM; CsWUS; cucumber; leaf development; shoot apical meristem..
© The Author 2015. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures



Similar articles
-
CsLFY is required for shoot meristem maintenance via interaction with WUSCHEL in cucumber (Cucumis sativus).New Phytol. 2018 Apr;218(1):344-356. doi: 10.1111/nph.14954. Epub 2017 Dec 23. New Phytol. 2018. PMID: 29274285
-
HANABA TARANU is a GATA transcription factor that regulates shoot apical meristem and flower development in Arabidopsis.Plant Cell. 2004 Oct;16(10):2586-600. doi: 10.1105/tpc.104.024869. Epub 2004 Sep 14. Plant Cell. 2004. PMID: 15367721 Free PMC article.
-
Transcription repressor HANABA TARANU controls flower development by integrating the actions of multiple hormones, floral organ specification genes, and GATA3 family genes in Arabidopsis.Plant Cell. 2013 Jan;25(1):83-101. doi: 10.1105/tpc.112.107854. Epub 2013 Jan 18. Plant Cell. 2013. PMID: 23335616 Free PMC article.
-
[HANABA TARANU, a GATA transcription factor which affects shoot apical meristem development].Seikagaku. 2006 Sep;78(9):888-91. Seikagaku. 2006. PMID: 17052027 Review. Japanese. No abstract available.
-
Molecular and Hormonal Regulation of Leaf Morphogenesis in Arabidopsis.Int J Mol Sci. 2020 Jul 20;21(14):5132. doi: 10.3390/ijms21145132. Int J Mol Sci. 2020. PMID: 32698541 Free PMC article. Review.
Cited by
-
Genetic regulation of shoot architecture in cucumber.Hortic Res. 2021 Jul 1;8(1):143. doi: 10.1038/s41438-021-00577-0. Hortic Res. 2021. PMID: 34193859 Free PMC article. Review.
-
Gibberellin biosynthesis is required for CPPU-induced parthenocarpy in melon.Hortic Res. 2023 May 3;10(6):uhad084. doi: 10.1093/hr/uhad084. eCollection 2023 Jun. Hortic Res. 2023. PMID: 37323228 Free PMC article.
-
CsBRC1 inhibits axillary bud outgrowth by directly repressing the auxin efflux carrier CsPIN3 in cucumber.Proc Natl Acad Sci U S A. 2019 Aug 20;116(34):17105-17114. doi: 10.1073/pnas.1907968116. Epub 2019 Aug 7. Proc Natl Acad Sci U S A. 2019. PMID: 31391306 Free PMC article.
-
Research Progress on the Leaf Morphology, Fruit Development and Plant Architecture of the Cucumber.Plants (Basel). 2022 Aug 16;11(16):2128. doi: 10.3390/plants11162128. Plants (Basel). 2022. PMID: 36015432 Free PMC article. Review.
-
Potential function of CbuSPL and gene encoding its interacting protein during flowering in Catalpa bungei.BMC Plant Biol. 2020 Mar 6;20(1):105. doi: 10.1186/s12870-020-2303-z. BMC Plant Biol. 2020. PMID: 32143577 Free PMC article.
References
-
- Aida M, Ishida T, Tasaka M. 1999. Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development 126, 1563–1570. - PubMed
-
- Aida M, Tasaka M. 2006. Genetic control of shoot organ boundaries. Current Opinion in Plant Biology 9, 72–77. - PubMed
-
- Atsmon D, Galun E. 1960. A morphogenetic study of staminate, pistillate and hermaphrodite flowers in Cucumis sativus L. Phytomorphology 10, 110–115.
-
- Bai S-L, Peng Y-B, Cui J-X, Gu H-T, Xu L-Y, Li Y-Q, Xu Z-H, Bai S-N. 2004. Developmental analyses reveal early arrests of the spore-bearing parts of reproductive organs in unisexual flowers of cucumber (Cucumis sativus L.). Planta 220, 230–240. - PubMed
-
- Bar M, Ori N. 2015. Compound leaf development in model plant species. Current Opinion in Plant Biology 23, 61–69. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

