Tissue-Specific Education of Decidual NK Cells
- PMID: 26320253
- PMCID: PMC4574523
- DOI: 10.4049/jimmunol.1501229
Tissue-Specific Education of Decidual NK Cells
Abstract
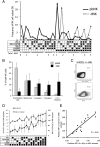
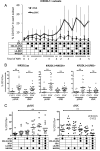
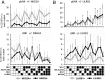
During human pregnancy, fetal trophoblast cells invade the decidua and remodel maternal spiral arteries to establish adequate nutrition during gestation. Tissue NK cells in the decidua (dNK) express inhibitory NK receptors (iNKR) that recognize allogeneic HLA-C molecules on trophoblast. Where this results in excessive dNK inhibition, the risk of pre-eclampsia or growth restriction is increased. However, the role of maternal, self-HLA-C in regulating dNK responsiveness is unknown. We investigated how the expression and function of five iNKR in dNK is influenced by maternal HLA-C. In dNK isolated from women who have HLA-C alleles that carry a C2 epitope, there is decreased expression frequency of the cognate receptor, KIR2DL1. In contrast, women with HLA-C alleles bearing a C1 epitope have increased frequency of the corresponding receptor, KIR2DL3. Maternal HLA-C had no significant effect on KIR2DL1 or KIR2DL3 in peripheral blood NK cells (pbNK). This resulted in a very different KIR repertoire for dNK capable of binding C1 or C2 epitopes compared with pbNK. We also show that, although maternal KIR2DL1 binding to C2 epitope educates dNK cells to acquire functional competence, the effects of other iNKR on dNK responsiveness are quite different from those in pbNK. This provides a basis for understanding how dNK responses to allogeneic trophoblast affect the outcome of pregnancy. Our findings suggest that the mechanisms that determine the repertoire of iNKR and the effect of self-MHC on NK education may differ in tissue NK cells compared with pbNK.
Copyright © 2015 The Authors.
Figures

References
-
- Raulet D. H., Vance R. E. 2006. Self-tolerance of natural killer cells. Nat. Rev. Immunol. 6: 520–531. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

