Codon Usage Influences the Local Rate of Translation Elongation to Regulate Co-translational Protein Folding
- PMID: 26321254
- PMCID: PMC4561030
- DOI: 10.1016/j.molcel.2015.07.018
Codon Usage Influences the Local Rate of Translation Elongation to Regulate Co-translational Protein Folding
Abstract
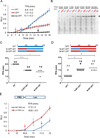
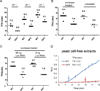
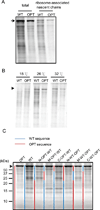
Codon usage bias is a universal feature of eukaryotic and prokaryotic genomes and has been proposed to regulate translation efficiency, accuracy, and protein folding based on the assumption that codon usage affects translation dynamics. The roles of codon usage in translation, however, are not clear and have been challenged by recent ribosome profiling studies. Here we used a Neurospora cell-free translation system to directly monitor the velocity of mRNA translation. We demonstrated that the preferred codons enhance the rate of translation elongation, whereas non-optimal codons slow elongation. Codon usage also controls ribosome traffic on mRNA. These conclusions were supported by ribosome profiling results in vitro and in vivo with template mRNAs designed to increase the signal-to-noise ratio. Finally, we demonstrate that codon usage regulates protein function by affecting co-translational protein folding. These results resolve a long-standing fundamental question and suggest the existence of a codon usage code for protein folding.
Copyright © 2015 Elsevier Inc. All rights reserved.
Conflict of interest statement
All authors declare that there is no financial conflict of interest that might be construed to influence the results or interpretation of our manuscript.
Figures




Comment in
-
Synthesis at the Speed of Codons.Trends Biochem Sci. 2015 Dec;40(12):717-718. doi: 10.1016/j.tibs.2015.10.005. Epub 2015 Oct 31. Trends Biochem Sci. 2015. PMID: 26526516
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

