HNRNPA2B1 Is a Mediator of m(6)A-Dependent Nuclear RNA Processing Events
- PMID: 26321680
- PMCID: PMC4673968
- DOI: 10.1016/j.cell.2015.08.011
HNRNPA2B1 Is a Mediator of m(6)A-Dependent Nuclear RNA Processing Events
Abstract
N(6)-methyladenosine (m(6)A) is the most abundant internal modification of messenger RNA. While the presence of m(6)A on transcripts can impact nuclear RNA fates, a reader of this mark that mediates processing of nuclear transcripts has not been identified. We find that the RNA-binding protein HNRNPA2B1 binds m(6)A-bearing RNAs in vivo and in vitro and its biochemical footprint matches the m(6)A consensus motif. HNRNPA2B1 directly binds a set of nuclear transcripts and elicits similar alternative splicing effects as the m(6)A writer METTL3. Moreover, HNRNPA2B1 binds to m(6)A marks in a subset of primary miRNA transcripts, interacts with the microRNA Microprocessor complex protein DGCR8, and promotes primary miRNA processing. Also, HNRNPA2B1 loss and METTL3 depletion cause similar processing defects for these pri-miRNA precursors. We propose HNRNPA2B1 to be a nuclear reader of the m(6)A mark and to mediate, in part, this mark's effects on primary microRNA processing and alternative splicing. PAPERCLIP.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures

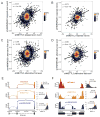
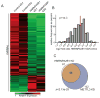
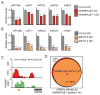


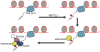
References
-
- Csepany T, Lin A, Baldick CJ, Jr, Beemon K. Sequence specificity of mRNA N6-adenosine methyltransferase. The Journal of biological chemistry. 1990;265:20117–20122. - PubMed
-
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

