MINEs: open access databases of computationally predicted enzyme promiscuity products for untargeted metabolomics
- PMID: 26322134
- PMCID: PMC4550642
- DOI: 10.1186/s13321-015-0087-1
MINEs: open access databases of computationally predicted enzyme promiscuity products for untargeted metabolomics
Abstract
Background: In spite of its great promise, metabolomics has proven difficult to execute in an untargeted and generalizable manner. Liquid chromatography-mass spectrometry (LC-MS) has made it possible to gather data on thousands of cellular metabolites. However, matching metabolites to their spectral features continues to be a bottleneck, meaning that much of the collected information remains uninterpreted and that new metabolites are seldom discovered in untargeted studies. These challenges require new approaches that consider compounds beyond those available in curated biochemistry databases.
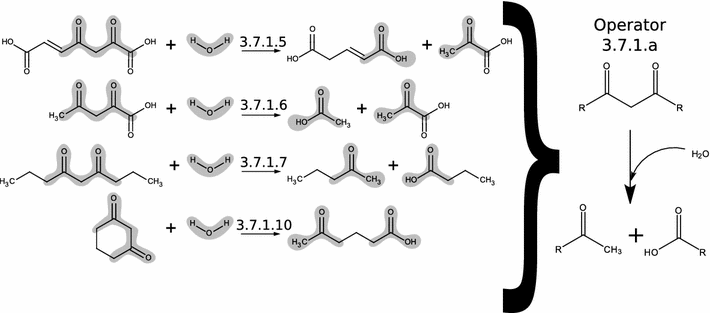
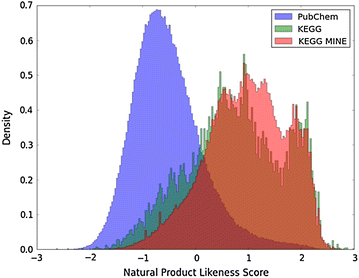
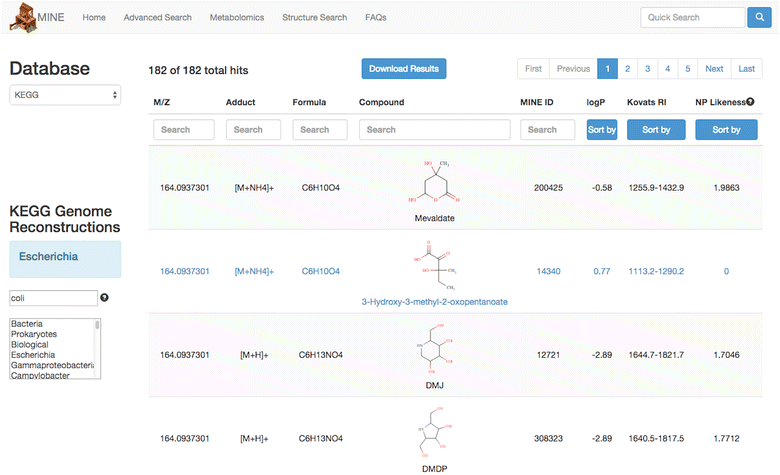
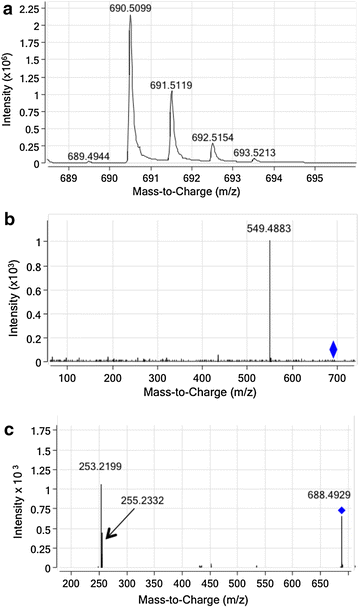
Description: Here we present Metabolic In silico Network Expansions (MINEs), an extension of known metabolite databases to include molecules that have not been observed, but are likely to occur based on known metabolites and common biochemical reactions. We utilize an algorithm called the Biochemical Network Integrated Computational Explorer (BNICE) and expert-curated reaction rules based on the Enzyme Commission classification system to propose the novel chemical structures and reactions that comprise MINE databases. Starting from the Kyoto Encyclopedia of Genes and Genomes (KEGG) COMPOUND database, the MINE contains over 571,000 compounds, of which 93% are not present in the PubChem database. However, these MINE compounds have on average higher structural similarity to natural products than compounds from KEGG or PubChem. MINE databases were able to propose annotations for 98.6% of a set of 667 MassBank spectra, 14% more than KEGG alone and equivalent to PubChem while returning far fewer candidates per spectra than PubChem (46 vs. 1715 median candidates). Application of MINEs to LC-MS accurate mass data enabled the identity of an unknown peak to be confidently predicted.
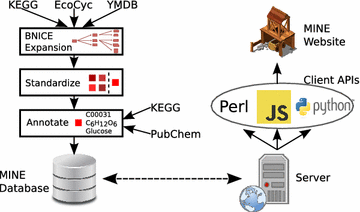
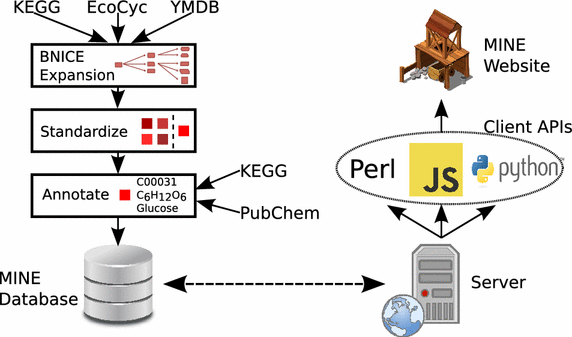
Conclusions: MINE databases are freely accessible for non-commercial use via user-friendly web-tools at http://minedatabase.mcs.anl.gov and developer-friendly APIs. MINEs improve metabolomics peak identification as compared to general chemical databases whose results include irrelevant synthetic compounds. Furthermore, MINEs complement and expand on previous in silico generated compound databases that focus on human metabolism. We are actively developing the database; future versions of this resource will incorporate transformation rules for spontaneous chemical reactions and more advanced filtering and prioritization of candidate structures. Graphical abstractMINE database construction and access methods. The process of constructing a MINE database from the curated source databases is depicted on the left. The methods for accessing the database are shown on the right.
Keywords: Enzyme promiscuity; Liquid chromatography–mass spectrometry; Metabolite identification; Untargeted metabolomics.
Figures
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

