Dopamine neurons projecting to the posterior striatum form an anatomically distinct subclass
- PMID: 26322384
- PMCID: PMC4598831
- DOI: 10.7554/eLife.10032
Dopamine neurons projecting to the posterior striatum form an anatomically distinct subclass
Abstract
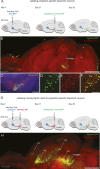
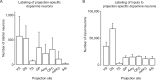
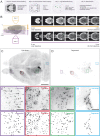
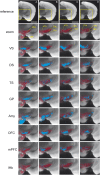
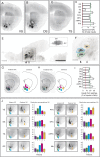
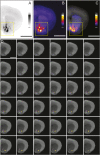
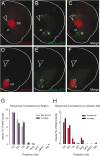
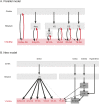
Combining rabies-virus tracing, optical clearing (CLARITY), and whole-brain light-sheet imaging, we mapped the monosynaptic inputs to midbrain dopamine neurons projecting to different targets (different parts of the striatum, cortex, amygdala, etc) in mice. We found that most populations of dopamine neurons receive a similar set of inputs rather than forming strong reciprocal connections with their target areas. A common feature among most populations of dopamine neurons was the existence of dense 'clusters' of inputs within the ventral striatum. However, we found that dopamine neurons projecting to the posterior striatum were outliers, receiving relatively few inputs from the ventral striatum and instead receiving more inputs from the globus pallidus, subthalamic nucleus, and zona incerta. These results lay a foundation for understanding the input/output structure of the midbrain dopamine circuit and demonstrate that dopamine neurons projecting to the posterior striatum constitute a unique class of dopamine neurons regulated by different inputs.
Keywords: anatomy; dopamine; input; monosynaptic; mouse; neuroscience; rabies virus; striatum.
Conflict of interest statement
JFB: Yoh Isogai and Joseph Bergan have filed a patent application on OptiView.
YI: Yoh Isogai and Joseph Bergan have filed a patent application on OptiView.
NU: Reviewing editor,
The other authors declare that no competing interests exist.
Figures





Comment in
-
Mapping neural circuits with CLARITY.Elife. 2015 Oct 9;4:e11409. doi: 10.7554/eLife.11409. Elife. 2015. PMID: 26452201 Free PMC article.
-
Selective connectivity of dopamine neurons projecting to the posterior striatum.Mov Disord. 2016 Mar;31(3):298. doi: 10.1002/mds.26496. Epub 2016 Feb 10. Mov Disord. 2016. PMID: 26861629 No abstract available.
References
-
- Backman CM, Malik N, Zhang Y, Shan L, Grinberg A, Hoffer BJ, Westphal H, Tomac AC. Characterization of a mouse strain expressing Cre recombinase from the 3′ untranslated region of the dopamine transporter locus. Genesis. 2006;44:383–390. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

