The DNA-binding inhibitor Id3 regulates IL-9 production in CD4(+) T cells
- PMID: 26322481
- PMCID: PMC5935106
- DOI: 10.1038/ni.3252
The DNA-binding inhibitor Id3 regulates IL-9 production in CD4(+) T cells
Abstract
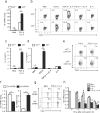
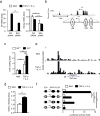
The molecular mechanisms by which signaling via transforming growth factor-β (TGF-β) and interleukin 4 (IL-4) control the differentiation of CD4(+) IL-9-producing helper T cells (TH9 cells) remain incompletely understood. We found here that the DNA-binding inhibitor Id3 regulated TH9 differentiation, as deletion of Id3 increased IL-9 production from CD4(+) T cells. Mechanistically, TGF-β1 and IL-4 downregulated Id3 expression, and this process required the kinase TAK1. A reduction in Id3 expression enhanced binding of the transcription factors E2A and GATA-3 to the Il9 promoter region, which promoted Il9 transcription. Notably, Id3-mediated control of TH9 differentiation regulated anti-tumor immunity in an experimental melanoma-bearing model in vivo and also in human CD4(+) T cells in vitro. Thus, our study reveals a previously unrecognized TAK1-Id3-E2A-GATA-3 pathway that regulates TH9 differentiation.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




References
-
- Schmitt E, Germann T, Goedert S, Hoehn P, Huels C, Koelsch S, et al. IL-9 production of naive CD4+ T cells depends on IL-2, is synergistically enhanced by a combination of TGF-beta and IL-4, and is inhibited by IFN-gamma. J Immunol. 1994;153(9):3989–3996. - PubMed
-
- Veldhoen M, Uyttenhove C, van Snick J, Helmby H, Westendorf A, Buer J, et al. Transforming growth factor-beta ‘reprograms’ the differentiation of T helper 2 cells and promotes an interleukin 9-producing subset. Nature immunology. 2008;9(12):1341–1346. - PubMed
-
- Cheng G, Arima M, Honda K, Hirata H, Eda F, Yoshida N, et al. Anti-interleukin-9 antibody treatment inhibits airway inflammation and hyperreactivity in mouse asthma model. American journal of respiratory and critical care medicine. 2002;166(3):409–416. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

