Lipid signalling couples translational surveillance to systemic detoxification in Caenorhabditis elegans
- PMID: 26322678
- PMCID: PMC4589496
- DOI: 10.1038/ncb3229
Lipid signalling couples translational surveillance to systemic detoxification in Caenorhabditis elegans
Erratum in
-
Author Correction: Lipid signalling couples translational surveillance to systemic detoxification in Caenorhabditis elegans.Nat Cell Biol. 2022 Jun;24(6):1005-1007. doi: 10.1038/s41556-022-00868-1. Nat Cell Biol. 2022. PMID: 35411089 No abstract available.
Abstract
Translation in eukaryotes is followed to detect toxins and virulence factors and coupled to the induction of defence pathways. Caenorhabditis elegans germline-specific mutations in translation components are detected by this system to induce detoxification and immune responses in distinct somatic cells. An RNA interference screen revealed gene inactivations that act at multiple steps in lipid biosynthetic and kinase pathways upstream of MAP kinase to mediate the systemic communication of translation defects to induce detoxification genes. Mammalian bile acids can rescue the defect in detoxification gene induction caused by C. elegans lipid biosynthetic gene inactivations. Extracts prepared from C. elegans with translation deficits but not from the wild type can also rescue detoxification gene induction in lipid-biosynthesis-defective strains. These eukaryotic antibacterial countermeasures are not ignored by bacteria: particular bacterial species suppress normal C. elegans detoxification responses to mutations in translation factors.
Conflict of interest statement
The authors declare no competing financial interests
Figures

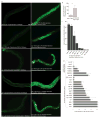
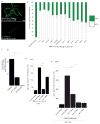
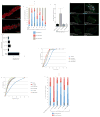
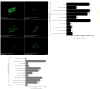

References
-
- Xu CC, Li CYTC, Kong ANTA. Induction of phase I, II and III drug metabolism/transport by xenobiotics. Arch Pharm Res. 2005;28:249–268. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

