Functional footprinting of regulatory DNA
- PMID: 26322838
- PMCID: PMC5381659
- DOI: 10.1038/nmeth.3554
Functional footprinting of regulatory DNA
Abstract
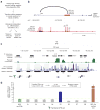
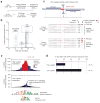
Regulatory regions harbor multiple transcription factor (TF) recognition sites; however, the contribution of individual sites to regulatory function remains challenging to define. We describe an approach that exploits the error-prone nature of genome editing-induced double-strand break repair to map functional elements within regulatory DNA at nucleotide resolution. We demonstrate the approach on a human erythroid enhancer, revealing single TF recognition sites that gate the majority of downstream regulatory function.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

