Microarray Analysis of Copy Number Variants on the Human Y Chromosome Reveals Novel and Frequent Duplications Overrepresented in Specific Haplogroups
- PMID: 26322892
- PMCID: PMC4554990
- DOI: 10.1371/journal.pone.0137223
Microarray Analysis of Copy Number Variants on the Human Y Chromosome Reveals Novel and Frequent Duplications Overrepresented in Specific Haplogroups
Abstract
Background: The human Y chromosome is almost always excluded from genome-wide investigations of copy number variants (CNVs) due to its highly repetitive structure. This chromosome should not be forgotten, not only for its well-known relevance in male fertility, but also for its involvement in clinical phenotypes such as cancers, heart failure and sex specific effects on brain and behaviour.
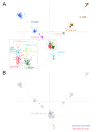
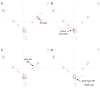
Results: We analysed Y chromosome data from Affymetrix 6.0 SNP arrays and found that the signal intensities for most of 8179 SNP/CN probes in the male specific region (MSY) discriminated between a male, background signals in a female and an isodicentric male containing a large deletion of the q-arm and a duplication of the p-arm of the Y chromosome. Therefore, this SNP/CN platform is suitable for identification of gain and loss of Y chromosome sequences. In a set of 1718 males, we found 25 different CNV patterns, many of which are novel. We confirmed some of these variants by PCR or qPCR. The total frequency of individuals with CNVs was 14.7%, including 9.5% with duplications, 4.5% with deletions and 0.7% exhibiting both. Hence, a novel observation is that the frequency of duplications was more than twice the frequency of deletions. Another striking result was that 10 of the 25 detected variants were significantly overrepresented in one or more haplogroups, demonstrating the importance to control for haplogroups in genome-wide investigations to avoid stratification. NO-M214(xM175) individuals presented the highest percentage (95%) of CNVs. If they were not counted, 12.4% of the rest included CNVs, and the difference between duplications (8.9%) and deletions (2.8%) was even larger.
Conclusions: Our results demonstrate that currently available genome-wide SNP platforms can be used to identify duplications and deletions in the human Y chromosome. Future association studies of the full spectrum of Y chromosome variants will demonstrate the potential involvement of gain or loss of Y chromosome sequence in different human phenotypes.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

