High-throughput optical sensing of nucleic acids in a nanopore array
- PMID: 26322943
- PMCID: PMC4821573
- DOI: 10.1038/nnano.2015.189
High-throughput optical sensing of nucleic acids in a nanopore array
Abstract
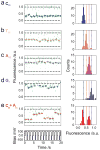
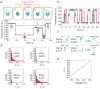
Protein nanopores such as α-haemolysin and Mycobacterium smegmatis porin A (MspA) can be used to sequence long strands of DNA at low cost. To provide high-speed sequencing, large arrays of nanopores are required, but current nanopore sequencing methods rely on ionic current measurements from individually addressed pores and such methods are likely to prove difficult to scale up. Here we show that, by optically encoding the ionic flux through protein nanopores, the discrimination of nucleic acid sequences and the detection of sequence-specific nucleic acid hybridization events can be parallelized. We make optical recordings at a density of ∼10(4) nanopores per mm(2) in a single droplet interface bilayer. Nanopore blockades can discriminate between DNAs with sub-picoampere equivalent resolution, and specific miRNA sequences can be identified by differences in unzipping kinetics. By creating an array of 2,500 bilayers with a micropatterned hydrogel chip, we are also able to load different samples into specific bilayers suitable for high-throughput nanopore recording.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

