Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index
- PMID: 26323059
- PMCID: PMC4589513
- DOI: 10.1038/ng.3390
Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index
Abstract
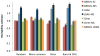
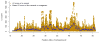
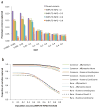
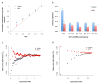
We propose a method (GREML-LDMS) to estimate heritability for human complex traits in unrelated individuals using whole-genome sequencing data. We demonstrate using simulations based on whole-genome sequencing data that ∼97% and ∼68% of variation at common and rare variants, respectively, can be captured by imputation. Using the GREML-LDMS method, we estimate from 44,126 unrelated individuals that all ∼17 million imputed variants explain 56% (standard error (s.e.) = 2.3%) of variance for height and 27% (s.e. = 2.5%) of variance for body mass index (BMI), and we find evidence that height- and BMI-associated variants have been under natural selection. Considering the imperfect tagging of imputation and potential overestimation of heritability from previous family-based studies, heritability is likely to be 60-70% for height and 30-40% for BMI. Therefore, the missing heritability is small for both traits. For further discovery of genes associated with complex traits, a study design with SNP arrays followed by imputation is more cost-effective than whole-genome sequencing at current prices.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

