Polycomb repressive complex PRC1 spatially constrains the mouse embryonic stem cell genome
- PMID: 26323060
- PMCID: PMC4847639
- DOI: 10.1038/ng.3393
Polycomb repressive complex PRC1 spatially constrains the mouse embryonic stem cell genome
Abstract
The Polycomb repressive complexes PRC1 and PRC2 maintain embryonic stem cell (ESC) pluripotency by silencing lineage-specifying developmental regulator genes. Emerging evidence suggests that Polycomb complexes act through controlling spatial genome organization. We show that PRC1 functions as a master regulator of mouse ESC genome architecture by organizing genes in three-dimensional interaction networks. The strongest spatial network is composed of the four Hox gene clusters and early developmental transcription factor genes, the majority of which contact poised enhancers. Removal of Polycomb repression leads to disruption of promoter-promoter contacts in the Hox gene network. In contrast, promoter-enhancer contacts are maintained in the absence of Polycomb repression, with accompanying widespread acquisition of active chromatin signatures at network enhancers and pronounced transcriptional upregulation of network genes. Thus, PRC1 physically constrains developmental transcription factor genes and their enhancers in a silenced but poised spatial network. We propose that the selective release of genes from this spatial network underlies cell fate specification during early embryonic development.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
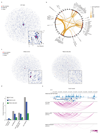


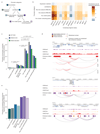

Comment in
-
PRC1 proteins orchestrate three-dimensional genome architecture.Nat Genet. 2015 Oct;47(10):1105-6. doi: 10.1038/ng.3411. Nat Genet. 2015. PMID: 26417860
References
Publication types
MeSH terms
Substances
Grants and funding
- BBS/E/B/0000H234/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 16358/CRUK_/Cancer Research UK/United Kingdom
- A16358/CRUK_/Cancer Research UK/United Kingdom
- BBS/E/B/000C0404/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

