Bunched and Madm Function Downstream of Tuberous Sclerosis Complex to Regulate the Growth of Intestinal Stem Cells in Drosophila
- PMID: 26323255
- PMCID: PMC4653243
- DOI: 10.1007/s12015-015-9617-5
Bunched and Madm Function Downstream of Tuberous Sclerosis Complex to Regulate the Growth of Intestinal Stem Cells in Drosophila
Abstract
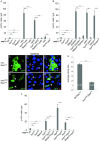
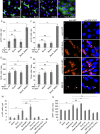
The Drosophila adult midgut contains intestinal stem cells that support homeostasis and repair. We show here that the leucine zipper protein Bunched and the adaptor protein Madm are novel regulators of intestinal stem cells. MARCM mutant clonal analysis and cell type specific RNAi revealed that Bunched and Madm were required within intestinal stem cells for proliferation. Transgenic expression of a tagged Bunched showed a cytoplasmic localization in midgut precursors, and the addition of a nuclear localization signal to Bunched reduced its function to cooperate with Madm to increase intestinal stem cell proliferation. Furthermore, the elevated cell growth and 4EBP phosphorylation phenotypes induced by loss of Tuberous Sclerosis Complex or overexpression of Rheb were suppressed by the loss of Bunched or Madm. Therefore, while the mammalian homolog of Bunched, TSC-22, is able to regulate transcription and suppress cancer cell proliferation, our data suggest the model that Bunched and Madm functionally interact with the TOR pathway in the cytoplasm to regulate the growth and subsequent division of intestinal stem cells.
Keywords: Bunched; Drosophila; Intestine; Madm; Stem cells; TSC-22; Tuberous sclerosis complex.
Figures


Similar articles
-
Non-canonical functions of the tuberous sclerosis complex-Rheb signalling axis.EMBO Mol Med. 2011 Apr;3(4):189-200. doi: 10.1002/emmm.201100131. Epub 2011 Mar 16. EMBO Mol Med. 2011. PMID: 21412983 Free PMC article. Review.
-
Tuberous sclerosis complex: from Drosophila to human disease.Trends Cell Biol. 2004 Feb;14(2):78-85. doi: 10.1016/j.tcb.2003.12.006. Trends Cell Biol. 2004. PMID: 15102439 Review.
-
Tuberous sclerosis complex and Myc coordinate the growth and division of Drosophila intestinal stem cells.J Cell Biol. 2011 May 16;193(4):695-710. doi: 10.1083/jcb.201103018. Epub 2011 May 9. J Cell Biol. 2011. PMID: 21555458 Free PMC article.
-
Drosophila TCTP is essential for growth and proliferation through regulation of dRheb GTPase.Nature. 2007 Feb 15;445(7129):785-8. doi: 10.1038/nature05528. Nature. 2007. PMID: 17301792
-
Madm (Mlf1 adapter molecule) cooperates with Bunched A to promote growth in Drosophila.J Biol. 2010;9(1):9. doi: 10.1186/jbiol216. Epub 2010 Feb 11. J Biol. 2010. PMID: 20149264 Free PMC article.
Cited by
-
Alternative splicing of bunched confers a dual role in hippo pathway-dependent growth and tumorigenesis.Oncogene. 2025 Jul;44(24):1949-1960. doi: 10.1038/s41388-025-03348-6. Epub 2025 Apr 2. Oncogene. 2025. PMID: 40175650
-
Basal stem cell progeny establish their apical surface in a junctional niche during turnover of an adult barrier epithelium.Nat Cell Biol. 2023 May;25(5):658-671. doi: 10.1038/s41556-023-01116-w. Epub 2023 Mar 30. Nat Cell Biol. 2023. PMID: 36997641 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- UL1TR000161/TR/NCATS NIH HHS/United States
- R01 GM060501/GM/NIGMS NIH HHS/United States
- UL1 TR000161/TR/NCATS NIH HHS/United States
- P30 DK032520/DK/NIDDK NIH HHS/United States
- P20 GM103440/GM/NIGMS NIH HHS/United States
- DK32520/DK/NIDDK NIH HHS/United States
- R01 DK083450/DK/NIDDK NIH HHS/United States
- R01 GM107457/GM/NIGMS NIH HHS/United States
- GM060501/GM/NIGMS NIH HHS/United States
- P20GM103440/GM/NIGMS NIH HHS/United States
- DK83450/DK/NIDDK NIH HHS/United States
- R01 GM105813/GM/NIGMS NIH HHS/United States
- GM105813/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

