Structure of RizA, an L-amino-acid ligase from Bacillus subtilis
- PMID: 26323296
- PMCID: PMC4555917
- DOI: 10.1107/S2053230X15012698
Structure of RizA, an L-amino-acid ligase from Bacillus subtilis
Abstract
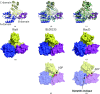
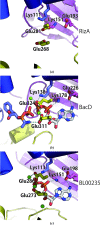
RizA is an L-amino-acid ligase from Bacillus subtilis that participates in the biosynthesis of rhizocticin, an oligopeptide antibiotic. The substrate-free form of RizA has been crystallized and the structure was solved at 2.8 Å resolution. The amino-acid-binding site appears to be capable of accommodating multiple amino acids, consistent with previous biochemical studies.
Keywords: ATP-grasp fold; dipeptide synthesis; l-amino-acid ligase; rhizocticin.
Figures


References
-
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
-
- Arai, T. & Kino, K. (2008). Biosci. Biotechnol. Biochem. 72, 3048–3050. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

