Hairpins under tension: RNA versus DNA
- PMID: 26323319
- PMCID: PMC4787782
- DOI: 10.1093/nar/gkv860
Hairpins under tension: RNA versus DNA
Abstract
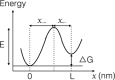
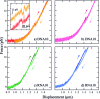
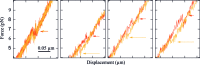
We use optical tweezers to control the folding and unfolding of individual DNA and RNA hairpins by force. Four hairpin molecules are studied in comparison: two DNA and two RNA ones. We observe that the conformational dynamics is slower for the RNA hairpins than for their DNA counterparts. Our results indicate that structures made of RNA are dynamically more stable. This difference might contribute to the fact that DNA and RNA play fundamentally different biological roles in spite of chemical similarity.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
-
- Bevilacqua P.C., Blose J.M. Structures, kinetics, thermodynamics, and biological functions of RNA hairpins. Annu. Rev. Phys. Chem. 2008;59:79–103. - PubMed
-
- Silverman S.K. Deoxyribozymes: DNA catalysts for bioorganic chemistry. Org. Biomol. Chem. 2004;2:2701–2706. - PubMed
-
- Astell C.R., Smith M., Chow M.B., Ward D.C. Structure of the 3′ hairpin termini of four rodent parvovirus genomes: Nucleotide sequence homologgy at origins of DNA replication. Cell. 1979;17:691–703. - PubMed
-
- Porschke D. Themodynamic and kinetic parameters of an oligonucleotide hairpin helix. Biophys. Chem. 1974;1:381–386. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

