PI(3,5)P2 controls endosomal branched actin dynamics by regulating cortactin-actin interactions
- PMID: 26323691
- PMCID: PMC4555817
- DOI: 10.1083/jcb.201412127
PI(3,5)P2 controls endosomal branched actin dynamics by regulating cortactin-actin interactions
Abstract
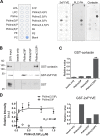
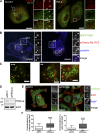
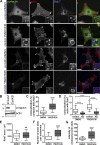
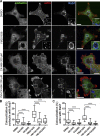
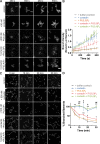
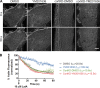
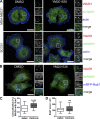
Branched actin critically contributes to membrane trafficking by regulating membrane curvature, dynamics, fission, and transport. However, how actin dynamics are controlled at membranes is poorly understood. Here, we identify the branched actin regulator cortactin as a direct binding partner of phosphatidylinositol 3,5-bisphosphate (PI(3,5)P2) and demonstrate that their interaction promotes turnover of late endosomal actin. In vitro biochemical studies indicated that cortactin binds PI(3,5)P2 via its actin filament-binding region. Furthermore, PI(3,5)P2 competed with actin filaments for binding to cortactin, thereby antagonizing cortactin activity. These findings suggest that PI(3,5)P2 formation on endosomes may remove cortactin from endosome-associated branched actin. Indeed, inhibition of PI(3,5)P2 production led to cortactin accumulation and actin stabilization on Rab7(+) endosomes. Conversely, inhibition of Arp2/3 complex activity greatly reduced cortactin localization to late endosomes. Knockdown of cortactin reversed PI(3,5)P2-inhibitor-induced actin accumulation and stabilization on endosomes. These data suggest a model in which PI(3,5)P2 binding removes cortactin from late endosomal branched actin networks and thereby promotes net actin turnover.
© 2015 Hong et al.
Figures


References
-
- Bird I.M. 1994. Analysis of cellular phosphoinositides and phosphoinositols by extraction and simple analytical procedures. Methods Mol. Biol. 27:227–248. - PubMed
-
- Bolino A., Muglia M., Conforti F.L., LeGuern E., Salih M.A., Georgiou D.M., Christodoulou K., Hausmanowa-Petrusewicz I., Mandich P., Schenone A., et al. 2000. Charcot-Marie-Tooth type 4B is caused by mutations in the gene encoding myotubularin-related protein-2. Nat. Genet. 25:17–19. 10.1038/75542 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- TR000445-06/TR/NCATS NIH HHS/United States
- UL1 TR000445/TR/NCATS NIH HHS/United States
- P60 DK020593/DK/NIDDK NIH HHS/United States
- R01 CA163592/CA/NCI NIH HHS/United States
- DK58404/DK/NIDDK NIH HHS/United States
- P30 DK058404/DK/NIDDK NIH HHS/United States
- R01GM075126/GM/NIGMS NIH HHS/United States
- P30 HD015052/HD/NICHD NIH HHS/United States
- R01 HL-55672/HL/NHLBI NIH HHS/United States
- EY08126/EY/NEI NIH HHS/United States
- P30 EY008126/EY/NEI NIH HHS/United States
- DK59637/DK/NIDDK NIH HHS/United States
- P30 DK020593/DK/NIDDK NIH HHS/United States
- R01 HL055672/HL/NHLBI NIH HHS/United States
- P30 CA068485/CA/NCI NIH HHS/United States
- CA68485/CA/NCI NIH HHS/United States
- R01 GM075126/GM/NIGMS NIH HHS/United States
- UL1 RR024975/RR/NCRR NIH HHS/United States
- R01CA163592/CA/NCI NIH HHS/United States
- HD15052/HD/NICHD NIH HHS/United States
- DK20593/DK/NIDDK NIH HHS/United States
- U24 DK059637/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

