Diversity and origins of anaerobic metabolism in mitochondria and related organelles
- PMID: 26323757
- PMCID: PMC4571565
- DOI: 10.1098/rstb.2014.0326
Diversity and origins of anaerobic metabolism in mitochondria and related organelles
Abstract
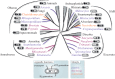
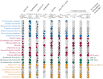
Across the diversity of life, organisms have evolved different strategies to thrive in hypoxic environments, and microbial eukaryotes (protists) are no exception. Protists that experience hypoxia often possess metabolically distinct mitochondria called mitochondrion-related organelles (MROs). While there are some common metabolic features shared between the MROs of distantly related protists, these organelles have evolved independently multiple times across the breadth of eukaryotic diversity. Until recently, much of our knowledge regarding the metabolic potential of different MROs was limited to studies in parasitic lineages. Over the past decade, deep-sequencing studies of free-living anaerobic protists have revealed novel configurations of metabolic pathways that have been co-opted for life in low oxygen environments. Here, we provide recent examples of anaerobic metabolism in the MROs of free-living protists and their parasitic relatives. Additionally, we outline evolutionary scenarios to explain the origins of these anaerobic pathways in eukaryotes.
Keywords: anaerobic metabolism; eukaryotic evolution; mitochondrion-related organelles.
© 2015 The Author(s).
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
