Horizontal gene flow from Eubacteria to Archaebacteria and what it means for our understanding of eukaryogenesis
- PMID: 26323767
- PMCID: PMC4571575
- DOI: 10.1098/rstb.2014.0337
Horizontal gene flow from Eubacteria to Archaebacteria and what it means for our understanding of eukaryogenesis
Abstract
The origin of the eukaryotic cell is considered one of the major evolutionary transitions in the history of life. Current evidence strongly supports a scenario of eukaryotic origin in which two prokaryotes, an archaebacterial host and an α-proteobacterium (the free-living ancestor of the mitochondrion), entered a stable symbiotic relationship. The establishment of this relationship was associated with a process of chimerization, whereby a large number of genes from the α-proteobacterial symbiont were transferred to the host nucleus. A general framework allowing the conceptualization of eukaryogenesis from a genomic perspective has long been lacking. Recent studies suggest that the origins of several archaebacterial phyla were coincident with massive imports of eubacterial genes. Although this does not indicate that these phyla originated through the same process that led to the origin of Eukaryota, it suggests that Archaebacteria might have had a general propensity to integrate into their genomes large amounts of eubacterial DNA. We suggest that this propensity provides a framework in which eukaryogenesis can be understood and studied in the light of archaebacterial ecology. We applied a recently developed supertree method to a genomic dataset composed of 392 eubacterial and 51 archaebacterial genera to test whether large numbers of genes flowing from Eubacteria are indeed coincident with the origin of major archaebacterial clades. In addition, we identified two potential large-scale transfers of uncertain directionality at the base of the archaebacterial tree. Our results are consistent with previous findings and seem to indicate that eubacterial gene imports (particularly from δ-Proteobacteria, Clostridia and Actinobacteria) were an important factor in archaebacterial history. Archaebacteria seem to have long relied on Eubacteria as a source of genetic diversity, and while the precise mechanism that allowed these imports is unknown, we suggest that our results support the view that processes comparable to those through which eukaryotes emerged might have been common in archaebacterial history.
Keywords: Archaebacteria; Bayesian supertrees; symbiosis; tree of life.
© 2015 The Author(s).
Figures
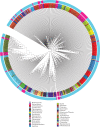

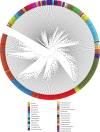
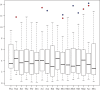

References
-
- Cavalier-Smith T. 2002. The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int. J. Syst. Evol. Microbiol. 52, 297–354. - PubMed
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
