Super-resolution mapping of glutamate receptors in C. elegans by confocal correlated PALM
- PMID: 26323790
- PMCID: PMC4555104
- DOI: 10.1038/srep13532
Super-resolution mapping of glutamate receptors in C. elegans by confocal correlated PALM
Erratum in
-
Corrigendum: Super-resolution mapping of glutamate receptors in C. elegans by confocal correlated PALM.Sci Rep. 2016 May 20;7:26047. doi: 10.1038/srep26047. Sci Rep. 2016. PMID: 27206048 Free PMC article. No abstract available.
Abstract
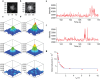
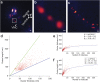
Photoactivated localization microscopy (PALM) is a super-resolution imaging technique based on the detection and subsequent localization of single fluorescent molecules. PALM is therefore a powerful tool in resolving structures and putative interactions of biomolecules at the ultimate analytical detection limit. However, its limited imaging depth restricts PALM mostly to in vitro applications. Considering the additional need for anatomical context when imaging a multicellular organism, these limitations render the use of PALM in whole animals difficult. Here we integrated PALM with confocal microscopy for correlated imaging of the C. elegans nervous system, a technique we termed confocal correlated PALM (ccPALM). The neurons, lying below several tissue layers, could be visualized up to 10 μm deep inside the animal. By ccPALM, we visualized ionotropic glutamate receptor distributions in C. elegans with an accuracy of 20 nm, revealing super-resolution structure of receptor clusters that we mapped onto annotated neurons in the animal. Pivotal to our results was the TIRF-independent detection of single molecules, achieved by genetic regulation of labeled receptor expression and localization to effectively reduce the background fluorescence. By correlating PALM with confocal microscopy, this platform enables dissecting biological structures with single molecule resolution in the physiologically relevant context of whole animals.
Figures


Similar articles
-
ExCel: Super-Resolution Imaging of C. elegans with Expansion Microscopy.Methods Mol Biol. 2022;2468:141-203. doi: 10.1007/978-1-0716-2181-3_9. Methods Mol Biol. 2022. PMID: 35320565 Free PMC article.
-
CemOrange2 fusions facilitate multifluorophore subcellular imaging in C. elegans.PLoS One. 2019 Mar 26;14(3):e0214257. doi: 10.1371/journal.pone.0214257. eCollection 2019. PLoS One. 2019. PMID: 30913273 Free PMC article.
-
Demonstrating Improved Multiple Transport-Mean-Free-Path Imaging Capabilities of Light Sheet Microscopy in the Quantification of Fluorescence Dynamics.Biotechnol J. 2018 Jan;13(1). doi: 10.1002/biot.201700419. Epub 2017 Dec 11. Biotechnol J. 2018. PMID: 29168308
-
Recent advances in super-resolution fluorescence imaging and its applications in biology.J Genet Genomics. 2013 Dec 20;40(12):583-95. doi: 10.1016/j.jgg.2013.11.003. Epub 2013 Nov 23. J Genet Genomics. 2013. PMID: 24377865 Review.
-
C. elegans cell cycle analysis.Methods Cell Biol. 2012;107:265-94. doi: 10.1016/B978-0-12-394620-1.00009-6. Methods Cell Biol. 2012. PMID: 22226527 Review.
Cited by
-
Impaired EAT-4 Vesicular Glutamate Transporter Leads to Defective Nocifensive Response of Caenorhabditis elegans to Noxious Heat.Neurochem Res. 2020 Apr;45(4):882-890. doi: 10.1007/s11064-020-02963-x. Epub 2020 Jan 16. Neurochem Res. 2020. PMID: 31950452
-
Organization and dynamics of the actin cytoskeleton during dendritic spine morphological remodeling.Cell Mol Life Sci. 2016 Aug;73(16):3053-73. doi: 10.1007/s00018-016-2214-1. Epub 2016 Apr 22. Cell Mol Life Sci. 2016. PMID: 27105623 Free PMC article. Review.
-
ExCel: Super-Resolution Imaging of C. elegans with Expansion Microscopy.Methods Mol Biol. 2022;2468:141-203. doi: 10.1007/978-1-0716-2181-3_9. Methods Mol Biol. 2022. PMID: 35320565 Free PMC article.
-
Digging deeper: methodologies for high-content phenotyping in Caenorhabditis elegans.Lab Anim (NY). 2019 Jul;48(7):207-216. doi: 10.1038/s41684-019-0326-6. Epub 2019 Jun 19. Lab Anim (NY). 2019. PMID: 31217565 Free PMC article. Review.
-
Expansion microscopy of C. elegans.Elife. 2020 May 1;9:e46249. doi: 10.7554/eLife.46249. Elife. 2020. PMID: 32356725 Free PMC article.
References
-
- Betzig E. et al. Imaging Intracellular Fluorescent Proteins at Nanometer Resolution. Science 313, 1642–1645 (2006). - PubMed
-
- Hell S. W. & Wichmann J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt. Lett. 19, 780–782 (1994). - PubMed
-
- Gustafsson M. G. L. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J Microsc 198, 82–87 (2000). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

