Spatial Interplay between Polycomb and Trithorax Complexes Controls Transcriptional Activity in T Lymphocytes
- PMID: 26324324
- PMCID: PMC4609743
- DOI: 10.1128/MCB.00677-15
Spatial Interplay between Polycomb and Trithorax Complexes Controls Transcriptional Activity in T Lymphocytes
Abstract
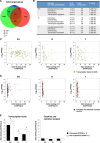
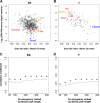
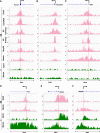
Trithorax group (TrxG) and Polycomb group (PcG) proteins are two mutually antagonistic chromatin modifying complexes, however, how they together mediate transcriptional counter-regulation remains unknown. Genome-wide analysis revealed that binding of Ezh2 and menin, central members of the PcG and TrxG complexes, respectively, were reciprocally correlated. Moreover, we identified a developmental change in the positioning of Ezh2 and menin in differentiated T lymphocytes compared to embryonic stem cells. Ezh2-binding upstream and menin-binding downstream of the transcription start site was frequently found at genes with higher transcriptional levels, and Ezh2-binding downstream and menin-binding upstream was found at genes with lower expression in T lymphocytes. Interestingly, of the Ezh2 and menin cooccupied genes, those exhibiting occupancy at the same position displayed greatly enhanced sensitivity to loss of Ezh2. Finally, we also found that different combinations of Ezh2 and menin occupancy were associated with expression of specific functional gene groups important for T cell development. Therefore, spatial cooperative gene regulation by the PcG and TrxG complexes may represent a novel mechanism regulating the transcriptional identity of differentiated cells.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures




Similar articles
-
The trithorax protein partner menin acts in tandem with EZH2 to suppress C/EBPα and differentiation in MLL-AF9 leukemia.Haematologica. 2013 Jun;98(6):918-27. doi: 10.3324/haematol.2012.074195. Epub 2013 Jan 24. Haematologica. 2013. PMID: 23349306 Free PMC article.
-
Menin regulates Inhbb expression through an Akt/Ezh2-mediated H3K27 histone modification.Biochim Biophys Acta Gene Regul Mech. 2017 Apr;1860(4):427-437. doi: 10.1016/j.bbagrm.2017.02.003. Epub 2017 Feb 12. Biochim Biophys Acta Gene Regul Mech. 2017. PMID: 28215965
-
Association of BMI1 with polycomb bodies is dynamic and requires PRC2/EZH2 and the maintenance DNA methyltransferase DNMT1.Mol Cell Biol. 2005 Dec;25(24):11047-58. doi: 10.1128/MCB.25.24.11047-11058.2005. Mol Cell Biol. 2005. PMID: 16314526 Free PMC article.
-
Genome Regulation by Polycomb and Trithorax: 70 Years and Counting.Cell. 2017 Sep 21;171(1):34-57. doi: 10.1016/j.cell.2017.08.002. Cell. 2017. PMID: 28938122 Review.
-
Therapeutic Opportunities of Targeting Canonical and Noncanonical PcG/TrxG Functions in Acute Myeloid Leukemia.Annu Rev Genomics Hum Genet. 2021 Aug 31;22:103-125. doi: 10.1146/annurev-genom-111120-102443. Epub 2021 Apr 30. Annu Rev Genomics Hum Genet. 2021. PMID: 33929894 Review.
Cited by
-
Epigenetic and Transcriptional Regulation in the Induction, Maintenance, Heterogeneity, and Recall-Response of Effector and Memory Th2 Cells.Front Immunol. 2018 Dec 12;9:2929. doi: 10.3389/fimmu.2018.02929. eCollection 2018. Front Immunol. 2018. PMID: 30619290 Free PMC article. Review.
-
CD4+ T-cell subsets in inflammatory diseases: beyond the Th1/Th2 paradigm.Int Immunol. 2016 Apr;28(4):163-71. doi: 10.1093/intimm/dxw006. Epub 2016 Feb 12. Int Immunol. 2016. PMID: 26874355 Free PMC article. Review.
-
Upregulation of RNA cap methyltransferase RNMT drives ribosome biogenesis during T cell activation.Nucleic Acids Res. 2021 Jul 9;49(12):6722-6738. doi: 10.1093/nar/gkab465. Nucleic Acids Res. 2021. PMID: 34125914 Free PMC article.
-
CD103hi Treg cells constrain lung fibrosis induced by CD103lo tissue-resident pathogenic CD4 T cells.Nat Immunol. 2019 Nov;20(11):1469-1480. doi: 10.1038/s41590-019-0494-y. Epub 2019 Oct 7. Nat Immunol. 2019. PMID: 31591568
-
Trichloroethylene-induced alterations in DNA methylation were enriched in polycomb protein binding sites in effector/memory CD4+ T cells.Environ Epigenet. 2017 Jul;3(3):dvx013. doi: 10.1093/eep/dvx013. Epub 2017 Sep 6. Environ Epigenet. 2017. PMID: 29129997 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
