Extreme Outlier Analysis Identifies Occult Mitogen-Activated Protein Kinase Pathway Mutations in Patients With Low-Grade Serous Ovarian Cancer
- PMID: 26324360
- PMCID: PMC4669594
- DOI: 10.1200/JCO.2015.62.4726
Extreme Outlier Analysis Identifies Occult Mitogen-Activated Protein Kinase Pathway Mutations in Patients With Low-Grade Serous Ovarian Cancer
Abstract
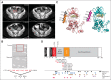
Purpose: No effective systemic therapy exists for patients with metastatic low-grade serous (LGS) ovarian cancers. BRAF and KRAS mutations are common in serous borderline (SB) and LGS ovarian cancers, and MEK inhibition has been shown to induce tumor regression in a minority of patients; however, no correlation has been observed between mutation status and clinical response. With the goal of identifying biomarkers of sensitivity to MEK inhibitor treatment, we performed an outlier analysis of a patient who experienced a complete, durable, and ongoing (> 5 years) response to selumetinib, a non-ATP competitive MEK inhibitor.
Patients and methods: Next-generation sequencing was used to analyze this patient's tumor as well as an additional 28 SB/LGS tumors. Functional characterization of an identified novel alteration of interest was performed.
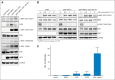
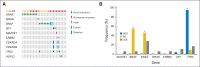
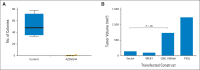
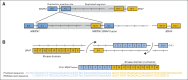
Results: Analysis of the extraordinary responder's tumor identified a 15-nucleotide deletion in the negative regulatory helix of the MAP2K1 gene encoding for MEK1. Functional characterization demonstrated that this mutant induced extracellular signal-regulated kinase pathway activation, promoted anchorage-independent growth and tumor formation in mice, and retained sensitivity to selumetinib. Analysis of additional LGS/SB tumors identified mutations predicted to induce extracellular signal-regulated kinase pathway activation in 82% (23 of 28), including two patients with BRAF fusions, one of whom achieved an ongoing complete response to MEK inhibitor-based combination therapy.
Conclusion: Alterations affecting the mitogen-activated protein kinase pathway are present in the majority of patients with LGS ovarian cancer. Next-generation sequencing analysis revealed deletions and fusions that are not detected by older sequencing approaches. These findings, coupled with the observation that a subset of patients with recurrent LGS ovarian cancer experienced dramatic and durable responses to MEK inhibitor therapy, support additional clinical studies of MEK inhibitors in this disease.
© 2015 by American Society of Clinical Oncology.
Conflict of interest statement
Authors' disclosures of potential conflicts of interest are found in the article online at
Figures
References
-
- Vang R, Shih IeM, Salani R, et al. Subdividing ovarian and peritoneal serous carcinoma into moderately differentiated and poorly differentiated does not have biologic validity based on molecular genetic and in vitro drug resistance data. Am J Surg Pathol. 2008;32:1667–1674. - PubMed
-
- Schmeler KM, Sun CC, Bodurka DC, et al. Neoadjuvant chemotherapy for low-grade serous carcinoma of the ovary or peritoneum. Gynecol Oncol. 2008;108:510–514. - PubMed
-
- Russell SE, McCluggage WG. A multistep model for ovarian tumorigenesis: The value of mutation analysis in the KRAS and BRAF genes. J Pathol. 2004;203:617–619. - PubMed
-
- Smith Sehdev AE, Sehdev PS, Kurman RJ. Noninvasive and invasive micropapillary (low-grade) serous carcinoma of the ovary: A clinicopathologic analysis of 135 cases. Am J Surg Pathol. 2003;27:725–736. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

