Transient bursts of Zscan4 expression are accompanied by the rapid derepression of heterochromatin in mouse embryonic stem cells
- PMID: 26324425
- PMCID: PMC4596397
- DOI: 10.1093/dnares/dsv013
Transient bursts of Zscan4 expression are accompanied by the rapid derepression of heterochromatin in mouse embryonic stem cells
Abstract
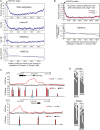
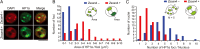
Mouse embryonic stem cells (mESCs) have a remarkable capacity to maintain normal genome stability and karyotype in culture. We previously showed that infrequent bursts of Zscan4 expression (Z4 events) are important for the maintenance of telomere length and genome stability in mESCs. However, the molecular details of Z4 events remain unclear. Here we show that Z4 events involve unexpected transcriptional derepression in heterochromatin regions that usually remain silent. During a Z4 event, we see rapid derepression and rerepression of heterochromatin leading to a burst of transcription that coincides with transient histone hyperacetylation and DNA demethylation, clustering of pericentromeric heterochromatin around the nucleolus, and accumulation of activating and repressive chromatin remodelling complexes. This heterochromatin-based transcriptional activity suggests that mESCs may maintain their extraordinary genome stability at least in part by transiently resetting their heterochromatin.
Keywords: embryonic stem cells; heterochromatin; pericentromere.
Published by Oxford University Press on behalf of Kazusa DNA Research Institute 2015. This work is written by (a) US Government employee(s) and is in the public domain in the US.
Figures





References
-
- Evans M.J., Kaufman M.H.. 1981, Establishment in culture of pluripotential cells from mouse embryos, Nature, 292, 154–6. - PubMed
-
- Bradley A., Evans M., Kaufman M.H., Robertson E.. 1984, Formation of germ-line chimaeras from embryo-derived teratocarcinoma cell lines, Nature, 309, 255–6. - PubMed
-
- Suda Y., Suzuki M., Ikawa Y., Aizawa S.. 1987, Mouse embryonic stem cells exhibit indefinite proliferative potential, J. Cell Physiol., 133, 197–201. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

