The Caulobacter crescentus Homolog of DnaA (HdaA) Also Regulates the Proteolysis of the Replication Initiator Protein DnaA
- PMID: 26324449
- PMCID: PMC4621095
- DOI: 10.1128/JB.00460-15
The Caulobacter crescentus Homolog of DnaA (HdaA) Also Regulates the Proteolysis of the Replication Initiator Protein DnaA
Abstract
It is not known how diverse bacteria regulate chromosome replication. Based on Escherichia coli studies, DnaA initiates replication and the homolog of DnaA (Hda) inactivates DnaA using the RIDA (regulatory inactivation of DnaA) mechanism that thereby prevents extra chromosome replication cycles. RIDA may be widespread, because the distantly related Caulobacter crescentus homolog HdaA also prevents extra chromosome replication (J. Collier and L. Shapiro, J Bacteriol 191:5706-5715, 2009, http://dx.doi.org/10.1128/JB.00525-09). To further study the HdaA/RIDA mechanism, we created a C. crescentus strain that shuts off hdaA transcription and rapidly clears HdaA protein. We confirm that HdaA prevents extra replication, since cells lacking HdaA accumulate extra chromosome DNA. DnaA binds nucleotides ATP and ADP, and our results are consistent with the established E. coli mechanism whereby Hda converts active DnaA-ATP to inactive DnaA-ADP. However, unlike E. coli DnaA, C. crescentus DnaA is also regulated by selective proteolysis. C. crescentus cells lacking HdaA reduce DnaA proteolysis in logarithmically growing cells, thereby implicating HdaA in this selective DnaA turnover mechanism. Also, wild-type C. crescentus cells remove all DnaA protein when they enter stationary phase. However, cells lacking HdaA retain stable DnaA protein even when they stop growing in nutrient-depleted medium that induces complete DnaA proteolysis in wild-type cells. Additional experiments argue for a distinct HdaA-dependent mechanism that selectively removes DnaA prior to stationary phase. Related freshwater Caulobacter species also remove DnaA during entry to stationary phase, implying a wider role for HdaA as a novel component of programed proteolysis.
Importance: Bacteria must regulate chromosome replication, and yet the mechanisms are not completely understood and not fully exploited for antibiotic development. Based on Escherichia coli studies, DnaA initiates replication, and the homolog of DnaA (Hda) inactivates DnaA to prevent extra replication. The distantly related Caulobacter crescentus homolog HdaA also regulates chromosome replication. Here we unexpectedly discovered that unlike the E. coli Hda, the C. crescentus HdaA also regulates DnaA proteolysis. Furthermore, this HdaA proteolysis acts in logarithmically growing and in stationary-phase cells and therefore in two very different physiological states. We argue that HdaA acts to help time chromosome replications in logarithmically growing cells and that it is an unexpected component of the programed entry into stationary phase.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
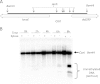


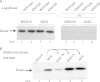

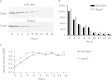


Similar articles
-
The β-sliding clamp directs the localization of HdaA to the replisome in Caulobacter crescentus.Microbiology (Reading). 2013 Nov;159(Pt 11):2237-2248. doi: 10.1099/mic.0.068577-0. Epub 2013 Aug 23. Microbiology (Reading). 2013. PMID: 23974073 Free PMC article.
-
Regulated degradation of chromosome replication proteins DnaA and CtrA in Caulobacter crescentus.Mol Microbiol. 2005 Feb;55(4):1233-45. doi: 10.1111/j.1365-2958.2004.04459.x. Mol Microbiol. 2005. PMID: 15686567
-
Feedback control of DnaA-mediated replication initiation by replisome-associated HdaA protein in Caulobacter.J Bacteriol. 2009 Sep;191(18):5706-16. doi: 10.1128/JB.00525-09. Epub 2009 Jul 24. J Bacteriol. 2009. PMID: 19633089 Free PMC article.
-
Regulation of the replication initiator DnaA in Caulobacter crescentus.Biochim Biophys Acta Gene Regul Mech. 2019 Jul;1862(7):697-705. doi: 10.1016/j.bbagrm.2018.01.004. Epub 2018 Jan 31. Biochim Biophys Acta Gene Regul Mech. 2019. PMID: 29382570 Review.
-
Regulation of chromosomal replication in Caulobacter crescentus.Plasmid. 2012 Mar;67(2):76-87. doi: 10.1016/j.plasmid.2011.12.007. Epub 2011 Dec 29. Plasmid. 2012. PMID: 22227374 Review.
Cited by
-
ClpAP is an auxiliary protease for DnaA degradation in Caulobacter crescentus.Mol Microbiol. 2016 Dec;102(6):1075-1085. doi: 10.1111/mmi.13537. Epub 2016 Oct 17. Mol Microbiol. 2016. PMID: 27667502 Free PMC article.
-
HdaB: a novel and conserved DnaA-related protein that targets the RIDA process to stimulate replication initiation.Nucleic Acids Res. 2020 Mar 18;48(5):2412-2423. doi: 10.1093/nar/gkz1193. Nucleic Acids Res. 2020. PMID: 31875223 Free PMC article.
-
Polyphosphate induces the proteolysis of ADP-bound fraction of initiator to inhibit DNA replication initiation upon stress in Escherichia coli.Nucleic Acids Res. 2020 Jun 4;48(10):5457-5466. doi: 10.1093/nar/gkaa217. Nucleic Acids Res. 2020. PMID: 32282902 Free PMC article.
-
Changing Perspectives on the Role of DnaA-ATP in Orisome Function and Timing Regulation.Front Microbiol. 2019 Aug 29;10:2009. doi: 10.3389/fmicb.2019.02009. eCollection 2019. Front Microbiol. 2019. PMID: 31555240 Free PMC article. Review.
-
Lon recognition of the replication initiator DnaA requires a bipartite degron.Mol Microbiol. 2019 Jan;111(1):176-186. doi: 10.1111/mmi.14146. Epub 2018 Nov 8. Mol Microbiol. 2019. PMID: 30288816 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

