An Extended Cyclic Di-GMP Network in the Predatory Bacterium Bdellovibrio bacteriovorus
- PMID: 26324450
- PMCID: PMC4686195
- DOI: 10.1128/JB.00422-15
An Extended Cyclic Di-GMP Network in the Predatory Bacterium Bdellovibrio bacteriovorus
Abstract
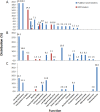
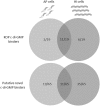
Over the course of the last 3 decades the role of the second messenger cyclic di-GMP (c-di-GMP) as a master regulator of bacterial physiology was determined. Although the control over c-di-GMP levels via synthesis and breakdown and the allosteric regulation of c-di-GMP over receptor proteins (effectors) and riboswitches have been extensively studied, relatively few effectors have been identified and most are of unknown functions. The obligate predatory bacterium Bdellovibrio bacteriovorus has a peculiar dimorphic life cycle, in which a phenotypic transition from a free-living attack phase (AP) to a sessile, intracellular predatory growth phase (GP) is tightly regulated by specific c-di-GMP diguanylate cyclases. B. bacteriovorus also bears one of the largest complement of defined effectors, almost none of known functions, suggesting that additional proteins may be involved in c-di-GMP signaling. In order to uncover novel c-di-GMP effectors, a c-di-GMP capture-compound mass-spectroscopy experiment was performed on wild-type AP and host-independent (HI) mutant cultures, the latter serving as a proxy for wild-type GP cells. Eighty-four proteins were identified as candidate c-di-GMP binders. Of these proteins, 65 did not include any recognized c-di-GMP binding site, and 3 carried known unorthodox binding sites. Putative functions could be assigned to 59 proteins. These proteins are included in metabolic pathways, regulatory circuits, cell transport, and motility, thereby creating a potentially large c-di-GMP network. False candidate effectors may include members of protein complexes, as well as proteins binding nucleotides or other cofactors that were, respectively, carried over or unspecifically interacted with the capture compound during the pulldown. Of the 84 candidates, 62 were found to specifically bind the c-di-GMP capture compound in AP or in HI cultures, suggesting c-di-GMP control over the whole-cell cycle of the bacterium. High affinity and specificity to c-di-GMP binding were confirmed using microscale thermophoresis with a hypothetical protein bearing a PilZ domain, an acyl coenzyme A dehydrogenase, and a two-component system response regulator, indicating that additional c-di-GMP binding candidates may be bona fide novel effectors.
Importance: In this study, 84 putative c-di-GMP binding proteins were identified in B. bacteriovorus, an obligate predatory bacterium whose lifestyle and reproduction are dependent on c-di-GMP signaling, using a c-di-GMP capture compound precipitation approach. This predicted complement covers metabolic, energy, transport, motility and regulatory pathways, and most of it is phase specific, i.e., 62 candidates bind the capture compound at defined modes of B. bacteriovorus lifestyle. Three of the putative binders further demonstrated specificity and high affinity to c-di-GMP via microscale thermophoresis, lending support for the presence of additional bona fide c-di-GMP effectors among the pulled-down protein repertoire.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures



References
-
- Hobley L, Fung RKY, Lambert C, Harris MATS, Dabhi JM, King SS, Basford SM, Uchida K, Till R, Ahmad R, Aizawa S-I, Gomelsky M, Sockett RE. 2012. Discrete cyclic di-GMP-dependent control of bacterial predation versus axenic growth in Bdellovibrio bacteriovorus. PLoS Pathog 8:e1002493. doi: 10.1371/journal.ppat.1002493. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

