Unique Role of the WD-40 Repeat Protein 5 (WDR5) Subunit within the Mixed Lineage Leukemia 3 (MLL3) Histone Methyltransferase Complex
- PMID: 26324722
- PMCID: PMC4646236
- DOI: 10.1074/jbc.M115.684142
Unique Role of the WD-40 Repeat Protein 5 (WDR5) Subunit within the Mixed Lineage Leukemia 3 (MLL3) Histone Methyltransferase Complex
Abstract
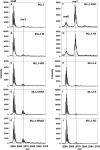
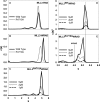
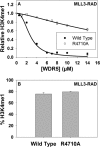
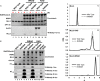
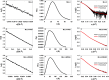
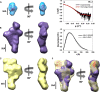
The MLL3 (mixed lineage leukemia 3) protein is a member of the human SET1 family of histone H3 lysine 4 methyltransferases and contains the conserved WDR5 interaction (Win) motif and the catalytic suppressor of variegation, enhancer of zeste, trithorax (SET) domain. The human SET1 family includes MLL1-4 and SETd1A/B, which all interact with a conserved subcomplex containing WDR5, RbBP5, Ash2L, and DPY-30 (WRAD) to form the minimal core complex required for full methyltransferase activity. However, recent evidence suggests that the WDR5 subunit may not be utilized in an identical manner within all SET1 family core complexes. Although the roles of WDR5 within the MLL1 core complex have been extensively studied, not much is known about the roles of WDR5 in other SET1 family core complexes. In this investigation, we set out to characterize the roles of the WDR5 subunit in the MLL3 core complex. We found that unlike MLL1, the MLL3 SET domain assembles with the RbBP5/Ash2L heterodimer independently of the Win motif-WDR5 interaction. Furthermore, we observed that WDR5 inhibits the monomethylation activity of the MLL3 core complex, which is dependent on the Win motif. We also found evidence suggesting that the WRAD subcomplex catalyzes weak H3K4 monomethylation within the context of the MLL3 core complex. Furthermore, solution structures of the MLL3 core complex assembled with and without WDR5 by small angle x-ray scattering show similar overall topologies. Together, this work demonstrates a unique role for WDR5 in modulating the enzymatic activity of the MLL3 core complex.
Keywords: chromatin; enzyme; histone; histone methylation; leukemia.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



References
-
- Milne T. A., Briggs S. D., Brock H. W., Martin M. E., Gibbs D., Allis C. D., and Hess J. L. (2002) MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol. Cell 10, 1107–1117 - PubMed
-
- Nakamura T., Mori T., Tada S., Krajewski W., Rozovskaia T., Wassell R., Dubois G., Mazo A., Croce C. M., and Canaani E. (2002) ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol. Cell 10, 1119–1128 - PubMed
-
- Dou Y., Milne T. A., Tackett A. J., Smith E. R., Fukuda A., Wysocka J., Allis C. D., Chait B. T., Hess J. L., and Roeder R. G. (2005) Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell 121, 873–885 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

