PLASTID MOVEMENT IMPAIRED1 and PLASTID MOVEMENT IMPAIRED1-RELATED1 Mediate Photorelocation Movements of Both Chloroplasts and Nuclei
- PMID: 26324877
- PMCID: PMC4587439
- DOI: 10.1104/pp.15.00214
PLASTID MOVEMENT IMPAIRED1 and PLASTID MOVEMENT IMPAIRED1-RELATED1 Mediate Photorelocation Movements of Both Chloroplasts and Nuclei
Abstract
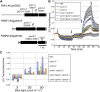
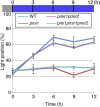
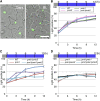
Organelle movement and positioning play important roles in fundamental cellular activities and adaptive responses to environmental stress in plants. To optimize photosynthetic light utilization, chloroplasts move toward weak blue light (the accumulation response) and escape from strong blue light (the avoidance response). Nuclei also move in response to strong blue light by utilizing the light-induced movement of attached plastids in leaf cells. Blue light receptor phototropins and several factors for chloroplast photorelocation movement have been identified through molecular genetic analysis of Arabidopsis (Arabidopsis thaliana). PLASTID MOVEMENT IMPAIRED1 (PMI1) is a plant-specific C2-domain protein that is required for efficient chloroplast photorelocation movement. There are two PLASTID MOVEMENT IMPAIRED1-RELATED (PMIR) genes, PMIR1 and PMIR2, in the Arabidopsis genome. However, the mechanism in which PMI1 regulates chloroplast and nuclear photorelocation movements and the involvement of PMIR1 and PMIR2 in these organelle movements remained unknown. Here, we analyzed chloroplast and nuclear photorelocation movements in mutant lines of PMI1, PMIR1, and PMIR2. In mesophyll cells, the pmi1 single mutant showed severe defects in both chloroplast and nuclear photorelocation movements resulting from the impaired regulation of chloroplast-actin filaments. In pavement cells, pmi1 mutant plants were partially defective in both plastid and nuclear photorelocation movements, but pmi1pmir1 and pmi1pmir1pmir2 mutant lines lacked the blue light-induced movement responses of plastids and nuclei completely. These results indicated that PMI1 is essential for chloroplast and nuclear photorelocation movements in mesophyll cells and that both PMI1 and PMIR1 are indispensable for photorelocation movements of plastids and thus, nuclei in pavement cells.
© 2015 American Society of Plant Biologists. All Rights Reserved.
Figures




References
-
- Castresana J. (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17: 540–552 - PubMed
-
- Cazzaniga S, Dall’ Osto L, Kong SG, Wada M, Bassi R (2013) Interaction between avoidance of photon absorption, excess energy dissipation and zeaxanthin synthesis against photooxidative stress in Arabidopsis. Plant J 76: 568–579 - PubMed
-
- Davis PA, Hangarter RP (2012) Chloroplast movement provides photoprotection to plants by redistributing PSII damage within leaves. Photosynth Res 112: 153–161 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

