Discovery of phosphonic acid natural products by mining the genomes of 10,000 actinomycetes
- PMID: 26324907
- PMCID: PMC4593130
- DOI: 10.1073/pnas.1500873112
Discovery of phosphonic acid natural products by mining the genomes of 10,000 actinomycetes
Abstract
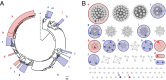
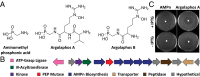
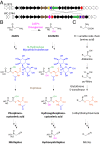
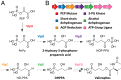
Although natural products have been a particularly rich source of human medicines, activity-based screening results in a very high rate of rediscovery of known molecules. Based on the large number of natural product biosynthetic genes in microbial genomes, many have proposed "genome mining" as an alternative approach for discovery efforts; however, this idea has yet to be performed experimentally on a large scale. Here, we demonstrate the feasibility of large-scale, high-throughput genome mining by screening a collection of over 10,000 actinomycetes for the genetic potential to make phosphonic acids, a class of natural products with diverse and useful bioactivities. Genome sequencing identified a diverse collection of phosphonate biosynthetic gene clusters within 278 strains. These clusters were classified into 64 distinct groups, of which 55 are likely to direct the synthesis of unknown compounds. Characterization of strains within five of these groups resulted in the discovery of a new archetypical pathway for phosphonate biosynthesis, the first (to our knowledge) dedicated pathway for H-phosphinates, and 11 previously undescribed phosphonic acid natural products. Among these compounds are argolaphos, a broad-spectrum antibacterial phosphonopeptide composed of aminomethylphosphonate in peptide linkage to a rare amino acid N(5)-hydroxyarginine; valinophos, an N-acetyl l-Val ester of 2,3-dihydroxypropylphosphonate; and phosphonocystoximate, an unusual thiohydroximate-containing molecule representing a new chemotype of sulfur-containing phosphonate natural products. Analysis of the genome sequences from the remaining strains suggests that the majority of the phosphonate biosynthetic repertoire of Actinobacteria has been captured at the gene level. This dereplicated strain collection now provides a reservoir of numerous, as yet undiscovered, phosphonate natural products.
Keywords: antibiotic; genome mining; natural products; phosphonic acid.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

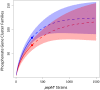
References
-
- Koehn FE, Carter GT. The evolving role of natural products in drug discovery. Nat Rev Drug Discov. 2005;4(3):206–220. - PubMed
-
- Brötz-Oesterhelt H, Sass P. Postgenomic strategies in antibacterial drug discovery. Future Microbiol. 2010;5(10):1553–1579. - PubMed
-
- Baltz RH. Marcel Faber Roundtable: Is our antibiotic pipeline unproductive because of starvation, constipation or lack of inspiration? J Ind Microbiol Biotechnol. 2006;33(7):507–513. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

