Arabidopsis ALIX is required for the endosomal localization of the deubiquitinating enzyme AMSH3
- PMID: 26324913
- PMCID: PMC4603487
- DOI: 10.1073/pnas.1510516112
Arabidopsis ALIX is required for the endosomal localization of the deubiquitinating enzyme AMSH3
Abstract
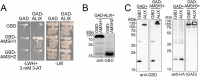
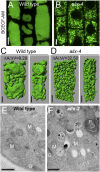
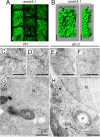
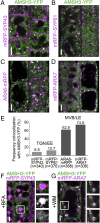
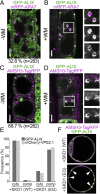
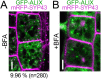
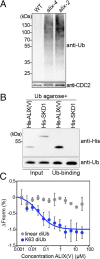
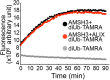
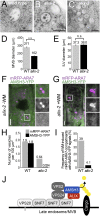
Ubiquitination is a signal for various cellular processes, including for endocytic degradation of plasma membrane cargos. Ubiquitinating as well as deubiquitinating enzymes (DUBs) can regulate these processes by modifying the ubiquitination status of target protein. Although accumulating evidence points to the important regulatory role of DUBs, the molecular basis of their regulation is still not well understood. Associated molecule with the SH3 domain of signal transduction adaptor molecule (STAM) (AMSH) is a conserved metalloprotease DUB in eukaryotes. AMSH proteins interact with components of the endosomal sorting complex required for transport (ESCRT) and are implicated in intracellular trafficking. To investigate how the function of AMSH is regulated at the cellular level, we carried out an interaction screen for the Arabidopsis AMSH proteins and identified the Arabidopsis homolog of apoptosis-linked gene-2 interacting protein X (ALIX) as a protein interacting with AMSH3 in vitro and in vivo. Analysis of alix knockout mutants in Arabidopsis showed that ALIX is essential for plant growth and development and that ALIX is important for the biogenesis of the vacuole and multivesicular bodies (MVBs). Cell biological analysis revealed that ALIX and AMSH3 colocalize on late endosomes. Although ALIX did not stimulate AMSH3 activity in vitro, in the absence of ALIX, AMSH3 localization on endosomes was abolished. Taken together, our data indicate that ALIX could function as an important regulator for AMSH3 function at the late endosomes.
Keywords: Arabidopsis; ESCRT-III; intracellular trafficking; ubiquitin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
The ESCRT-III-interacting deubiquitinating enzyme AMSH3 is essential for degradation of ubiquitinated membrane proteins in Arabidopsis thaliana.Plant Cell Physiol. 2014 Apr;55(4):727-36. doi: 10.1093/pcp/pcu019. Epub 2014 Jan 30. Plant Cell Physiol. 2014. PMID: 24486765
-
The Arabidopsis deubiquitinating enzyme AMSH3 interacts with ESCRT-III subunits and regulates their localization.Plant Cell. 2011 Aug;23(8):3026-40. doi: 10.1105/tpc.111.087254. Epub 2011 Aug 2. Plant Cell. 2011. PMID: 21810997 Free PMC article.
-
Arabidopsis SH3P2 is an ubiquitin-binding protein that functions together with ESCRT-I and the deubiquitylating enzyme AMSH3.Proc Natl Acad Sci U S A. 2017 Aug 22;114(34):E7197-E7204. doi: 10.1073/pnas.1710866114. Epub 2017 Aug 7. Proc Natl Acad Sci U S A. 2017. PMID: 28784794 Free PMC article.
-
Do Alix and ALG-2 really control endosomes for better or for worse?Biol Cell. 2006 Jan;98(1):69-77. doi: 10.1042/BC20050007. Biol Cell. 2006. PMID: 16354163 Review.
-
Plant endosomal trafficking pathways.Curr Opin Plant Biol. 2011 Dec;14(6):666-73. doi: 10.1016/j.pbi.2011.07.009. Epub 2011 Aug 5. Curr Opin Plant Biol. 2011. PMID: 21821464 Review.
Cited by
-
Brassinosteroid Signaling Dynamics: Ubiquitination-Dependent Regulation of Core Signaling Components.Int J Mol Sci. 2025 May 8;26(10):4502. doi: 10.3390/ijms26104502. Int J Mol Sci. 2025. PMID: 40429648 Free PMC article. Review.
-
Protein degrons and degradation: Exploring substrate recognition and pathway selection in plants.Plant Cell. 2024 Sep 3;36(9):3074-3098. doi: 10.1093/plcell/koae141. Plant Cell. 2024. PMID: 38701343 Free PMC article. Review.
-
Arabidopsis CaLB1 undergoes phase separation with the ESCRT protein ALIX and modulates autophagosome maturation.Nat Commun. 2024 Jun 19;15(1):5188. doi: 10.1038/s41467-024-49485-6. Nat Commun. 2024. PMID: 38898014 Free PMC article.
-
Genome-wide association study reveals new loci for yield-related traits in Sichuan wheat germplasm under stripe rust stress.BMC Genomics. 2019 Aug 8;20(1):640. doi: 10.1186/s12864-019-6005-6. BMC Genomics. 2019. PMID: 31395029 Free PMC article.
-
The Multivesicular Body and Autophagosome Pathways in Plants.Front Plant Sci. 2018 Dec 12;9:1837. doi: 10.3389/fpls.2018.01837. eCollection 2018. Front Plant Sci. 2018. PMID: 30619408 Free PMC article.
References
-
- Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. - PubMed
-
- Vierstra RD. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol. 2009;10(6):385–397. - PubMed
-
- Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. - PubMed
-
- Platta HW, Stenmark H. Endocytosis and signaling. Curr Opin Cell Biol. 2011;23(4):393–403. - PubMed
-
- Haglund K, Dikic I. The role of ubiquitylation in receptor endocytosis and endosomal sorting. J Cell Sci. 2012;125(Pt 2):265–275. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

