PopZ identifies the new pole, and PodJ identifies the old pole during polar growth in Agrobacterium tumefaciens
- PMID: 26324921
- PMCID: PMC4577194
- DOI: 10.1073/pnas.1515544112
PopZ identifies the new pole, and PodJ identifies the old pole during polar growth in Agrobacterium tumefaciens
Abstract
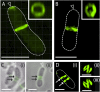
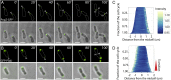
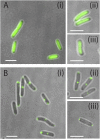
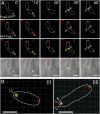
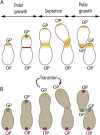
Agrobacterium tumefaciens elongates by addition of peptidoglycan (PG) only at the pole created by cell division, the growth pole, whereas the opposite pole, the old pole, is inactive for PG synthesis. How Agrobacterium assigns and maintains pole asymmetry is not understood. Here, we investigated whether polar growth is correlated with novel pole-specific localization of proteins implicated in a variety of growth and cell division pathways. The cell cycle of A. tumefaciens was monitored by time-lapse and superresolution microscopy to image the localization of A. tumefaciens homologs of proteins involved in cell division, PG synthesis and pole identity. FtsZ and FtsA accumulate at the growth pole during elongation, and improved imaging reveals FtsZ disappears from the growth pole and accumulates at the midcell before FtsA. The L,D-transpeptidase Atu0845 was detected mainly at the growth pole. A. tumefaciens specific pole-organizing protein (Pop) PopZAt and polar organelle development (Pod) protein PodJAt exhibited dynamic yet distinct behavior. PopZAt was found exclusively at the growing pole and quickly switches to the new growth poles of both siblings immediately after septation. PodJAt is initially at the old pole but then also accumulates at the growth pole as the cell cycle progresses suggesting that PodJAt may mediate the transition of the growth pole to an old pole. Thus, PopZAt is a marker for growth pole identity, whereas PodJAt identifies the old pole.
Keywords: Agrobacterium cell cycle; PodJ; PopZ; bacterial cell division; polar growth.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Loss of PodJ in Agrobacterium tumefaciens Leads to Ectopic Polar Growth, Branching, and Reduced Cell Division.J Bacteriol. 2016 Jun 13;198(13):1883-1891. doi: 10.1128/JB.00198-16. Print 2016 Jul 1. J Bacteriol. 2016. PMID: 27137498 Free PMC article.
-
Loss of PopZ At activity in Agrobacterium tumefaciens by Deletion or Depletion Leads to Multiple Growth Poles, Minicells, and Growth Defects.mBio. 2017 Nov 14;8(6):e01881-17. doi: 10.1128/mBio.01881-17. mBio. 2017. PMID: 29138309 Free PMC article.
-
Absence of the Polar Organizing Protein PopZ Results in Reduced and Asymmetric Cell Division in Agrobacterium tumefaciens.J Bacteriol. 2017 Aug 8;199(17):e00101-17. doi: 10.1128/JB.00101-17. Print 2017 Sep 1. J Bacteriol. 2017. PMID: 28630123 Free PMC article.
-
The essential features and modes of bacterial polar growth.Trends Microbiol. 2015 Jun;23(6):347-53. doi: 10.1016/j.tim.2015.01.003. Epub 2015 Feb 3. Trends Microbiol. 2015. PMID: 25662291 Review.
-
Recent advances on the development of bacterial poles.Trends Microbiol. 2004 Nov;12(11):518-25. doi: 10.1016/j.tim.2004.09.003. Trends Microbiol. 2004. PMID: 15488393 Review.
Cited by
-
Segregation of four Agrobacterium tumefaciens replicons during polar growth: PopZ and PodJ control segregation of essential replicons.Proc Natl Acad Sci U S A. 2020 Oct 20;117(42):26366-26373. doi: 10.1073/pnas.2014371117. Epub 2020 Oct 6. Proc Natl Acad Sci U S A. 2020. PMID: 33024016 Free PMC article.
-
Polar Organizing Protein PopZ Is Required for Chromosome Segregation in Agrobacterium tumefaciens.J Bacteriol. 2017 Aug 8;199(17):e00111-17. doi: 10.1128/JB.00111-17. Print 2017 Sep 1. J Bacteriol. 2017. PMID: 28630129 Free PMC article.
-
RdsA Is a Global Regulator That Controls Cell Shape and Division in Rhizobium etli.Front Microbiol. 2022 Apr 7;13:858440. doi: 10.3389/fmicb.2022.858440. eCollection 2022. Front Microbiol. 2022. PMID: 35464952 Free PMC article.
-
Hit the right spots: cell cycle control by phosphorylated guanosines in alphaproteobacteria.Nat Rev Microbiol. 2017 Mar;15(3):137-148. doi: 10.1038/nrmicro.2016.183. Epub 2017 Jan 31. Nat Rev Microbiol. 2017. PMID: 28138140 Review.
-
MapB, the Brucella suis TamB homologue, is involved in cell envelope biogenesis, cell division and virulence.Sci Rep. 2019 Feb 15;9(1):2158. doi: 10.1038/s41598-018-37668-3. Sci Rep. 2019. PMID: 30770847 Free PMC article.
References
-
- Zupan J, Muth TR, Draper O, Zambryski P. The transfer of DNA from agrobacterium tumefaciens into plants: A feast of fundamental insights. Plant J. 2000;23(1):11–28. - PubMed
-
- den Blaauwen T, de Pedro MA, Nguyen-Distèche M, Ayala JA. Morphogenesis of rod-shaped sacculi. FEMS Microbiol Rev. 2008;32(2):321–344. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

