Balancing between affinity and speed in target DNA search by zinc-finger proteins via modulation of dynamic conformational ensemble
- PMID: 26324943
- PMCID: PMC4577168
- DOI: 10.1073/pnas.1507726112
Balancing between affinity and speed in target DNA search by zinc-finger proteins via modulation of dynamic conformational ensemble
Abstract
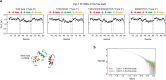
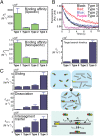
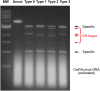
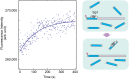
Although engineering of transcription factors and DNA-modifying enzymes has drawn substantial attention for artificial gene regulation and genome editing, most efforts focus on affinity and specificity of the DNA-binding proteins, typically overlooking the kinetic properties of these proteins. However, a simplistic pursuit of high affinity can lead to kinetically deficient proteins that spend too much time at nonspecific sites before reaching their targets on DNA. We demonstrate that structural dynamic knowledge of the DNA-scanning process allows for kinetically and thermodynamically balanced engineering of DNA-binding proteins. Our current study of the zinc-finger protein Egr-1 (also known as Zif268) and its nuclease derivatives reveals kinetic and thermodynamic roles of the dynamic conformational equilibrium between two modes during the DNA-scanning process: one mode suitable for search and the other for recognition. By mutagenesis, we were able to shift this equilibrium, as confirmed by NMR spectroscopy. Using fluorescence and biochemical assays as well as computational simulations, we analyzed how the shifts of the conformational equilibrium influence binding affinity, target search kinetics, and efficiency in displacing other proteins from the target sites. A shift toward the recognition mode caused an increase in affinity for DNA and a decrease in search efficiency. In contrast, a shift toward the search mode caused a decrease in affinity and an increase in search efficiency. This accelerated site-specific DNA cleavage by the zinc-finger nuclease, without enhancing off-target cleavage. Our study shows that appropriate modulation of the dynamic conformational ensemble can greatly improve zinc-finger technology, which has used Egr-1 (Zif268) as a major scaffold for engineering.
Keywords: DNA scanning; dynamics; kinetics; protein–DNA interactions; target search.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







Similar articles
-
Towards understanding the molecular recognition process in prokaryotic zinc-finger domain.Eur J Med Chem. 2015 Feb 16;91:100-8. doi: 10.1016/j.ejmech.2014.09.040. Epub 2014 Sep 16. Eur J Med Chem. 2015. PMID: 25240418
-
Interaction identification of Zif268 and TATA(ZF) proteins with GC-/AT-rich DNA sequence: A theoretical study.J Comput Chem. 2011 Feb;32(3):416-28. doi: 10.1002/jcc.21630. J Comput Chem. 2011. PMID: 20658568
-
Solution structure of the first three zinc fingers of TFIIIA bound to the cognate DNA sequence: determinants of affinity and sequence specificity.J Mol Biol. 1997 Oct 17;273(1):183-206. doi: 10.1006/jmbi.1997.1291. J Mol Biol. 1997. PMID: 9367756
-
Artificial zinc finger DNA binding domains: versatile tools for genome engineering and modulation of gene expression.J Cell Biochem. 2015 Nov;116(11):2435-44. doi: 10.1002/jcb.25226. J Cell Biochem. 2015. PMID: 25989233 Free PMC article. Review.
-
Keep your fingers off my DNA: protein-protein interactions mediated by C2H2 zinc finger domains.Cell Biochem Biophys. 2008;50(3):111-31. doi: 10.1007/s12013-008-9008-5. Epub 2008 Feb 6. Cell Biochem Biophys. 2008. PMID: 18253864 Review.
Cited by
-
Poly(ADP-ribose) polymerase 1 searches DNA via a 'monkey bar' mechanism.Elife. 2018 Aug 8;7:e37818. doi: 10.7554/eLife.37818. Elife. 2018. PMID: 30088474 Free PMC article.
-
Importance of Binding Affinity for the Activity of a Metallodendritic Chemical Nuclease.Pharmaceutics. 2018 Dec 3;10(4):258. doi: 10.3390/pharmaceutics10040258. Pharmaceutics. 2018. PMID: 30513860 Free PMC article.
-
Facilitated diffusion of Argonaute-mediated target search.RNA Biol. 2019 Sep;16(9):1093-1107. doi: 10.1080/15476286.2019.1616353. Epub 2019 May 20. RNA Biol. 2019. PMID: 31068066 Free PMC article. Review.
-
Electrostatic Interactions in Protein Structure, Folding, Binding, and Condensation.Chem Rev. 2018 Feb 28;118(4):1691-1741. doi: 10.1021/acs.chemrev.7b00305. Epub 2018 Jan 10. Chem Rev. 2018. PMID: 29319301 Free PMC article. Review.
-
Discrete-state stochastic kinetic models for target DNA search by proteins: Theory and experimental applications.Biophys Chem. 2021 Feb;269:106521. doi: 10.1016/j.bpc.2020.106521. Epub 2020 Dec 10. Biophys Chem. 2021. PMID: 33338872 Free PMC article. Review.
References
-
- Choo Y, Isalan M. Advances in zinc finger engineering. Curr Opin Struct Biol. 2000;10(4):411–416. - PubMed
-
- Pabo CO, Peisach E, Grant RA. Design and selection of novel Cys2His2 zinc finger proteins. Annu Rev Biochem. 2001;70:313–340. - PubMed
-
- Segal DJ, Barbas CF., 3rd Custom DNA-binding proteins come of age: Polydactyl zinc-finger proteins. Curr Opin Biotechnol. 2001;12(6):632–637. - PubMed
-
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11(9):636–646. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

