Discovery of a Novel Immune Gene Signature with Profound Prognostic Value in Colorectal Cancer: A Model of Cooperativity Disorientation Created in the Process from Development to Cancer
- PMID: 26325386
- PMCID: PMC4556644
- DOI: 10.1371/journal.pone.0137171
Discovery of a Novel Immune Gene Signature with Profound Prognostic Value in Colorectal Cancer: A Model of Cooperativity Disorientation Created in the Process from Development to Cancer
Abstract
Immune response-related genes play a major role in colorectal carcinogenesis by mediating inflammation or immune-surveillance evasion. Although remarkable progress has been made to investigate the underlying mechanism, the understanding of the complicated carcinogenesis process was enormously hindered by large-scale tumor heterogeneity. Development and carcinogenesis share striking similarities in their cellular behavior and underlying molecular mechanisms. The association between embryonic development and carcinogenesis makes embryonic development a viable reference model for studying cancer thereby circumventing the potentially misleading complexity of tumor heterogeneity. Here we proposed that the immune genes, responsible for intra-immune cooperativity disorientation (defined in this study as disruption of developmental expression correlation patterns during carcinogenesis), probably contain untapped prognostic resource of colorectal cancer. In this study, we determined the mRNA expression profile of 137 human biopsy samples, including samples from different stages of human colonic development, colorectal precancerous progression and colorectal cancer samples, among which 60 were also used to generate miRNA expression profile. We originally established Spearman correlation transition model to quantify the cooperativity disorientation associated with the transition from normal to precancerous to cancer tissue, in conjunction with miRNA-mRNA regulatory network and machine learning algorithm to identify genes with prognostic value. Finally, a 12-gene signature was extracted, whose prognostic value was evaluated using Kaplan-Meier survival analysis in five independent datasets. Using the log-rank test, the 12-gene signature was closely related to overall survival in four datasets (GSE17536, n = 177, p = 0.0054; GSE17537, n = 55, p = 0.0039; GSE39582, n = 562, p = 0.13; GSE39084, n = 70, p = 0.11), and significantly associated with disease-free survival in four datasets (GSE17536, n = 177, p = 0.0018; GSE17537, n = 55, p = 0.016; GSE39582, n = 557, p = 4.4e-05; GSE14333, n = 226, p = 0.032). Cox regression analysis confirmed that the 12-gene signature was an independent factor in predicting colorectal cancer patient's overall survival (hazard ratio: 1.759; 95% confidence interval: 1.126-2.746; p = 0.013], as well as disease-free survival (hazard ratio: 2.116; 95% confidence interval: 1.324-3.380; p = 0.002).
Conflict of interest statement
Figures

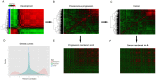

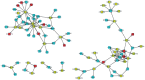
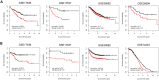
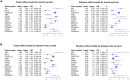
References
-
- Troiani T, Martinelli E, Napolitano S, Morgillo F, Belli G, Cioffi L, et al. Molecular aspects of resistance to biological and non-biological drugs and strategies to overcome resistance in colorectal cancer. Curr Med Chem. 2014;21(14):1639–53. Epub 2013/09/03. . - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

