Genomic and Phenomic Study of Mammary Pathogenic Escherichia coli
- PMID: 26327312
- PMCID: PMC4556653
- DOI: 10.1371/journal.pone.0136387
Genomic and Phenomic Study of Mammary Pathogenic Escherichia coli
Abstract
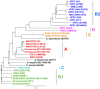
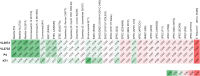
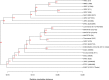
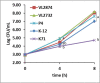
Escherichia coli is a major etiological agent of intra-mammary infections (IMI) in cows, leading to acute mastitis and causing great economic losses in dairy production worldwide. Particular strains cause persistent IMI, leading to recurrent mastitis. Virulence factors of mammary pathogenic E. coli (MPEC) involved pathogenesis of mastitis as well as those differentiating strains causing acute or persistent mastitis are largely unknown. This study aimed to identify virulence markers in MPEC through whole genome and phenome comparative analysis. MPEC strains causing acute (VL2874 and P4) or persistent (VL2732) mastitis were compared to an environmental strain (K71) and to the genomes of strains representing different E. coli pathotypes. Intra-mammary challenge in mice confirmed experimentally that the strains studied here have different pathogenic potential, and that the environmental strain K71 is non-pathogenic in the mammary gland. Analysis of whole genome sequences and predicted proteomes revealed high similarity among MPEC, whereas MPEC significantly differed from the non-mammary pathogenic strain K71, and from E. coli genomes from other pathotypes. Functional features identified in MPEC genomes and lacking in the non-mammary pathogenic strain were associated with synthesis of lipopolysaccharide and other membrane antigens, ferric-dicitrate iron acquisition and sugars metabolism. Features associated with cytotoxicity or intra-cellular survival were found specifically in the genomes of strains from severe and acute (VL2874) or persistent (VL2732) mastitis, respectively. MPEC genomes were relatively similar to strain K-12, which was subsequently shown here to be possibly pathogenic in the mammary gland. Phenome analysis showed that the persistent MPEC was the most versatile in terms of nutrients metabolized and acute MPEC the least. Among phenotypes unique to MPEC compared to the non-mammary pathogenic strain were uric acid and D-serine metabolism. This study reveals virulence factors and phenotypic characteristics of MPEC that may play a role in pathogenesis of E. coli mastitis.
Conflict of interest statement
Figures
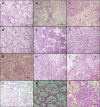
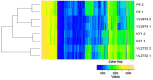
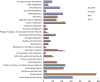
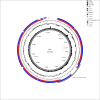

References
-
- Halasa T, Nielen M, Huirne RBM, Hogeveen H (2009) Stochastic bio-economic model of bovine intramammary infection. Livest Sci 124: 295–305. Available: http://www.sciencedirect.com/science/article/pii/S1871141309000651. Accessed 12 May 2014.
-
- Hogeveen H, Huijps K, Lam TJGM (2011) Economic aspects of mastitis: new developments. N Z Vet J 59: 16–23. Available: http://www.ncbi.nlm.nih.gov/pubmed/21328153. Accessed 17 September 2014. 10.1080/00480169.2011.547165 - DOI - PubMed
-
- Le Maréchal C, Thiéry R, Vautor E, Le Loir Y (2011) Mastitis impact on technological properties of milk and quality of milk products-A review. Dairy Sci Technol 91: 247–282. Available: http://link.springer.com/10.1007/s13594-011-0009-6. Accessed 12 May 2014. - DOI
-
- Leitner G, Merin U, Silanikove N (2011) Effects of glandular bacterial infection and stage of lactation on milk clotting parameters: Comparison among cows, goats and sheep. Int Dairy J 21: 279–285. Available: http://www.sciencedirect.com/science/article/pii/S095869461000261X. Accessed 18 October 2014.
-
- Bradley AJ (2002) Bovine Mastitis: An Evolving Disease. Vet J 164: 116–128. Available: http://www.sciencedirect.com/science/article/pii/S1090023302907240. Accessed 3 June 2014. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- SRA/SRP059659
- SRA/SRP059660
- SRA/SRP059661
- SRA/SRP059662
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

