A Yeast Mutant Deleted of GPH1 Bears Defects in Lipid Metabolism
- PMID: 26327557
- PMCID: PMC4556709
- DOI: 10.1371/journal.pone.0136957
A Yeast Mutant Deleted of GPH1 Bears Defects in Lipid Metabolism
Abstract
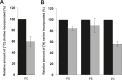
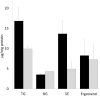
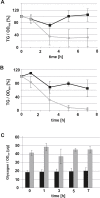
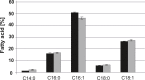
In a previous study we demonstrated up-regulation of the yeast GPH1 gene under conditions of phosphatidylethanolamine (PE) depletion caused by deletion of the mitochondrial (M) phosphatidylserine decarboxylase 1 (PSD1) (Gsell et al., 2013, PLoS One. 8(10):e77380. doi: 10.1371/journal.pone.0077380). Gph1p has originally been identified as a glycogen phosphorylase catalyzing degradation of glycogen to glucose in the stationary growth phase of the yeast. Here we show that deletion of this gene also causes decreased levels of phosphatidylcholine (PC), triacylglycerols and steryl esters. Depletion of the two non-polar lipids in a Δgph1 strain leads to lack of lipid droplets, and decrease of the PC level results in instability of the plasma membrane. In vivo labeling experiments revealed that formation of PC via both pathways of biosynthesis, the cytidine diphosphate (CDP)-choline and the methylation route, is negatively affected by a Δgph1 mutation, although expression of genes involved is not down regulated. Altogether, Gph1p besides its function as a glycogen mobilizing enzyme appears to play a regulatory role in yeast lipid metabolism.
Conflict of interest statement
Figures





References
-
- Athenstaedt K, Daum G. Tgl4p and Tgl5p, two triacylglycerol lipases of the yeast Saccharomyces cerevisiae are localized to lipid particles J. Biol. Chem. 2005; 280: 37301–37309. - PubMed
-
- Daum G, Lees ND, Bard M, Dickson R. Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae , Yeast. 1998; 14: 1471–1510. - PubMed
-
- Carman GM, Henry SA. Phospholipid biosynthesis in the yeast Saccharomyces cerevisiae and interrelationship with other metabolic processes, Prog Lipid Res. 1999; 38: 361–399. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

