Paradoxical resistance of multiple myeloma to proteasome inhibitors by decreased levels of 19S proteasomal subunits
- PMID: 26327694
- PMCID: PMC4602331
- DOI: 10.7554/eLife.08153
Paradoxical resistance of multiple myeloma to proteasome inhibitors by decreased levels of 19S proteasomal subunits
Abstract
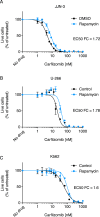
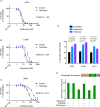
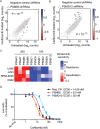
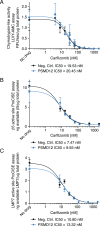
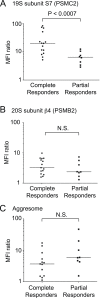
Hallmarks of cancer, including rapid growth and aneuploidy, can result in non-oncogene addiction to the proteostasis network that can be exploited clinically. The defining example is the exquisite sensitivity of multiple myeloma (MM) to 20S proteasome inhibitors, such as carfilzomib. However, MM patients invariably acquire resistance to these drugs. Using a next-generation shRNA platform, we found that proteostasis factors, including chaperones and stress-response regulators, controlled the response to carfilzomib. Paradoxically, 19S proteasome regulator knockdown induced resistance to carfilzomib in MM and non-MM cells. 19S subunit knockdown did not affect the activity of the 20S subunits targeted by carfilzomib nor their inhibition by the drug, suggesting an alternative mechanism, such as the selective accumulation of protective factors. In MM patients, lower 19S levels predicted a diminished response to carfilzomib-based therapies. Together, our findings suggest that an understanding of network rewiring can inform development of new combination therapies to overcome drug resistance.
Trial registration: ClinicalTrials.gov NCT01402284.
Keywords: cancer; carfilzomib; cell biology; human; myeloma; proteasome; proteostasis.
Conflict of interest statement
TJB: is an employee of Onyx Pharmaceuticals, an Amgen subsidiary.
AGL: is an employee of Onyx Pharmaceuticals, an Amgen subsidiary.
The authors declare that no competing interests exist.
Figures







References
-
- Bianchi G, Oliva L, Cascio P, Pengo N, Fontana F, Cerruti F, Orsi A, Pasqualetto E, Mezghrani A, Calbi V, Palladini G, Giuliani N, Anderson KC, Sitia R, Cenci S. The proteasome load versus capacity balance determines apoptotic sensitivity of multiple myeloma cells to proteasome inhibition. Blood. 2009;113:3040–3049. doi: 10.1182/blood-2008-08-172734. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

