Immunohistochemical, genetic and epigenetic profiles of hereditary and triple negative breast cancers. Relevance in personalized medicine
- PMID: 26328265
- PMCID: PMC4548346
Immunohistochemical, genetic and epigenetic profiles of hereditary and triple negative breast cancers. Relevance in personalized medicine
Abstract
This study aims to identify the profile of immunohistochemical (IHC) parameters, copy number aberrations (CNAs) and epigenetic alterations [promoter methylation (PM) and miR expression] related to hereditary (H) and triple negative (TN) breast cancer (BC). This profile could be of relevance for guiding tumor response to treatment with targeting therapy. The study comprises 278 formalin fixed paraffin-embedded BCs divided into two groups: H group, including 88 hereditary BC (HBC) and 190 non hereditary (NHBC), and TN group, containing 79 TNBC and 187 non TNBC (NTNBC). We assessed IHC parameters (Ki67, ER, PR, HER2, CK5/6, CK18 and Cadherin-E), CNA of 20 BC related genes, and PM of 24 tumor suppressor genes employing MLPA/MS-MLPA (MRC Holland, Amsterdam). MiR-4417, miR-423-3p, miR-590-5p and miR-187-3p expression was assessed by quantitative RT-PCR (Applied Biosystems). Binary logistic regression was applied to select the parameters that better differentiate the HBC or TN groups. For HBC we found that, ER expression, ERBB2 CNA and PM in RASSF1 and TIMP3 were associated with NHBC whereas; MYC and AURKA CNA were linked to HBC. For TNBC, we found that CDC6 CNA, GSTP1 and RASSF1 PM and miR-423-3p hyperexpression were characteristic of NTNBC, while MYC aberrations, BRCA1 hypermethylation and miR-590-5p and miR-4417 hyperexpression were more indicative of TNBC. The selected markers allow establishing BC subtypes, which are characterized by showing similar etiopathogenetic mechanisms, some of them being molecular targets for known drugs or possible molecular targets. These results could be the basis to implement a personalized therapy.
Keywords: BRCA1; BRCA2; Sporadic breast cancer; hereditary breast cancer; miR expression profile; molecular markers; mutations.
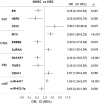
Figures


References
-
- Boyle P, Ferlay J. Cancer incidence and mortality in Europe. Ann Oncol. 2004;16:481–488. - PubMed
-
- Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W, Bell R, Rosenthal J, Hussey C, Tran T, McClure M, Frye C, Hattier T, Phelps R, Haugen-Strano A, Katcher H, Yakumo K, Gholami Z, Shaffer D, Stone S, Bayer S, Wray C, Bogden R, Dayananth P, Ward J, Tonin P, Narod S, Bristow PK, Norris FH, Helvering L, Morrison P, Rosteck P, Lai M, Barrett JC, Lewis C, Neuhausen S, Albright L, Goldgar D, Wiseman R, Kamb A, Skolnick MH. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. - PubMed
-
- Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C, Micklem G. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–792. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
