Large scale transcriptome analysis reveals interplay between development of forest trees and a beneficial mycorrhiza helper bacterium
- PMID: 26328611
- PMCID: PMC4557895
- DOI: 10.1186/s12864-015-1856-y
Large scale transcriptome analysis reveals interplay between development of forest trees and a beneficial mycorrhiza helper bacterium
Abstract
Background: Pedunculate oak, Quercus robur is an abundant forest tree species that hosts a large and diverse community of beneficial ectomycorrhizal fungi (EMFs), whereby ectomycorrhiza (EM) formation is stimulated by mycorrhiza helper bacteria such as Streptomyces sp. AcH 505. Oaks typically grow rhythmically, with alternating root flushes (RFs) and shoot flushes (SFs). We explored the poorly understood mechanisms by which oaks integrate signals induced by their beneficial microbes and endogenous rhythmic growth at the level of gene expression. To this end, we compared transcript profiles of oak microcuttings at RF and SF during interactions with AcH 505 alone and in combination with the basidiomycetous EMF Piloderma croceum.
Results: The local root and distal leaf responses to the microorganisms differed substantially. More genes involved in the recognition of bacteria and fungi, defence and cell wall remodelling related transcription factors (TFs) were differentially expressed in the roots than in the leaves of oaks. In addition, interaction with AcH 505 and P. croceum affected the expression of a higher number of genes during SF than during RF, including AcH 505 elicited defence response, which was attenuated by co-inoculation with P. croceum in the roots during SF. Genes encoding leucine-rich receptor-like kinases (LRR-RLKs) and proteins (LRR-RLPs), LRR containing defence response regulators, TFs from bZIP, ERF and WRKY families, xyloglucan cell wall transglycolases/hydrolases and exordium proteins were differentially expressed in both roots and leaves of plants treated with AcH 505. Only few genes, including specific RLKs and TFs, were induced in both AcH 505 and co-inoculation treatments.
Conclusion: Treatment with AcH 505 induces and maintains the expression levels of signalling genes encoding candidate receptor protein kinases and TFs and leads to differential expression of cell wall modification related genes in pedunculate oak microcuttings. Local gene expression response to AcH 505 alone and in combination with P. croceum are more pronounced when roots are in resting stages, possibly due to the fact that non growing roots re-direct their activity towards plant defence rather than growth.
Figures
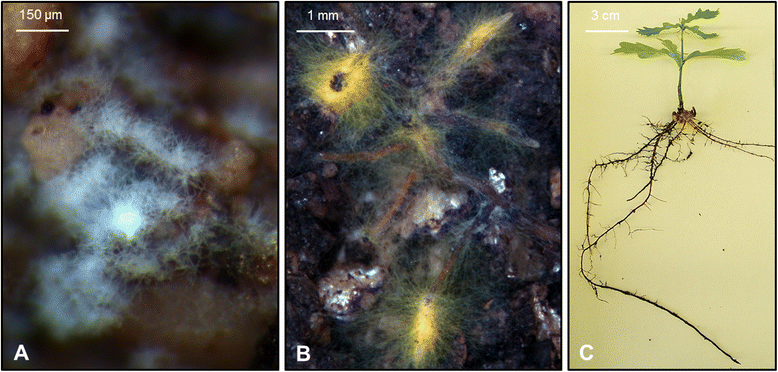
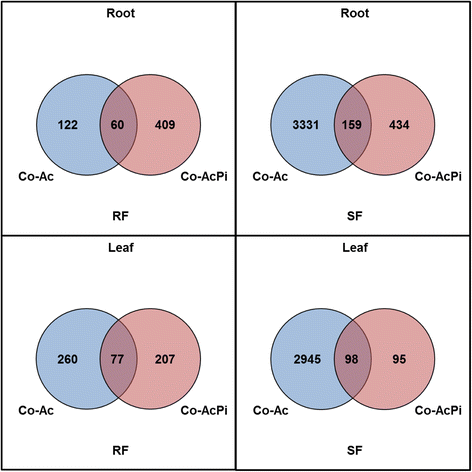


References
-
- Smith SE, Read DJ. Mycorrhizal symbiosis. London: Academic; 2008.
-
- Riedlinger J, Schrey SD, Tarkka MT, Hampp R, Kapur M, Fiedler HP. Auxofuran, a novel metabolite that stimulates the growth of fly agaric, is produced by the mycorrhiza helper bacterium Streptomyces strain AcH 505. Appl Environ Microbiol. 2006;72(5):3550–3557. doi: 10.1128/AEM.72.5.3550-3557.2006. - DOI - PMC - PubMed
-
- Brulé C, Frey-Klett P, Pierrat JC, Courrier S, Gerard F, Lemoine MC, Rousselet JL, Sommer G, Garbaye J. Survival in the soil of the ectomycorrhizal fungus Laccaria bicolor and the effects of a mycorrhiza helper Pseudomonas fluorescens. Soil Biol Biochem. 2001;33(12–13):1683–1694. doi: 10.1016/S0038-0717(01)00090-6. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

