Analysis methods for studying the 3D architecture of the genome
- PMID: 26328929
- PMCID: PMC4556012
- DOI: 10.1186/s13059-015-0745-7
Analysis methods for studying the 3D architecture of the genome
Abstract
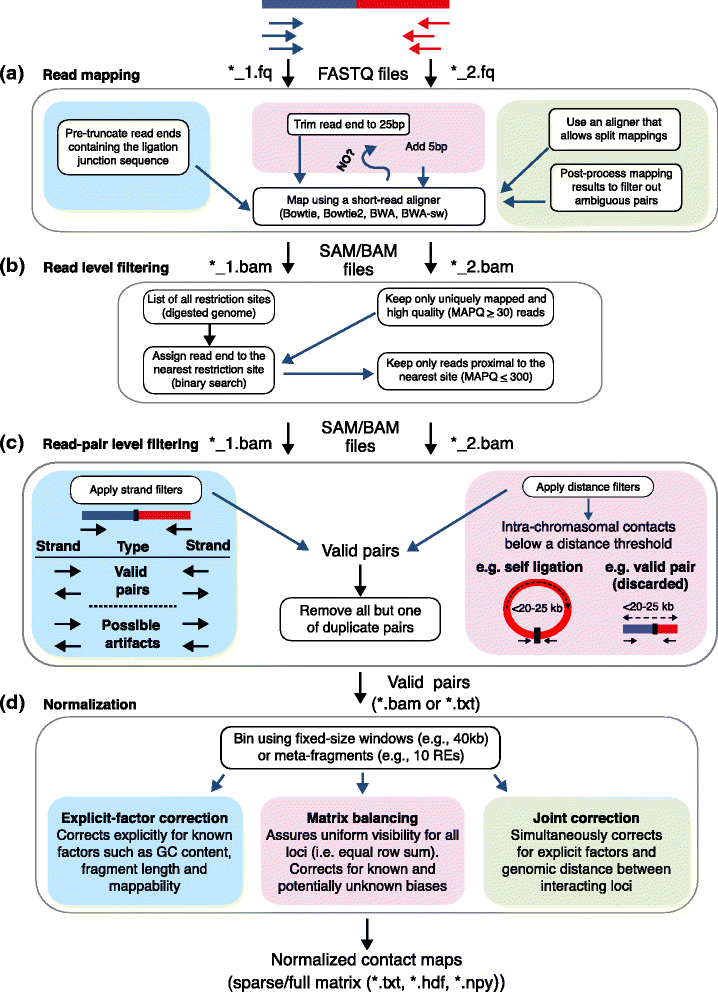
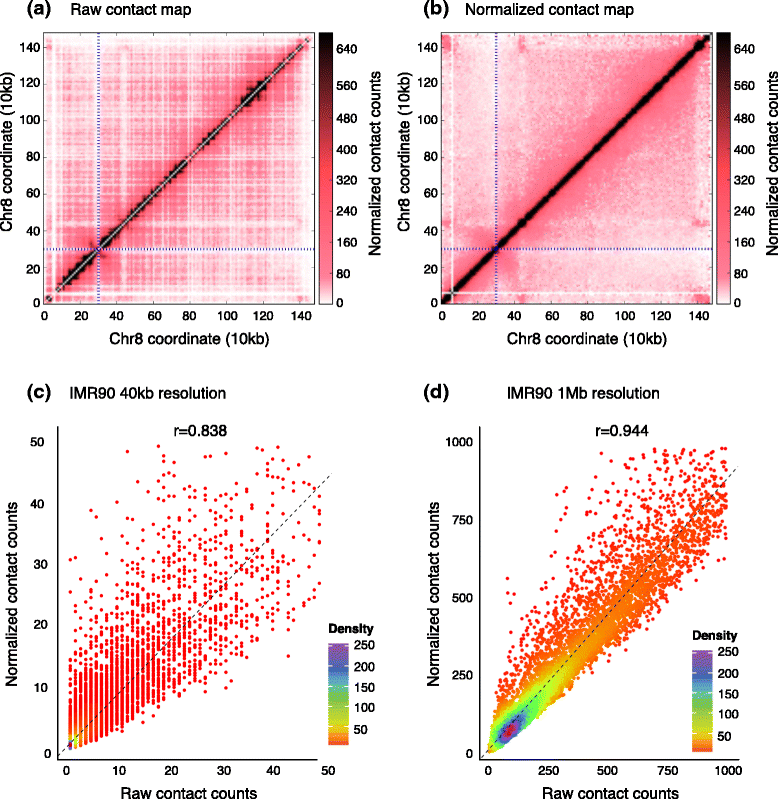
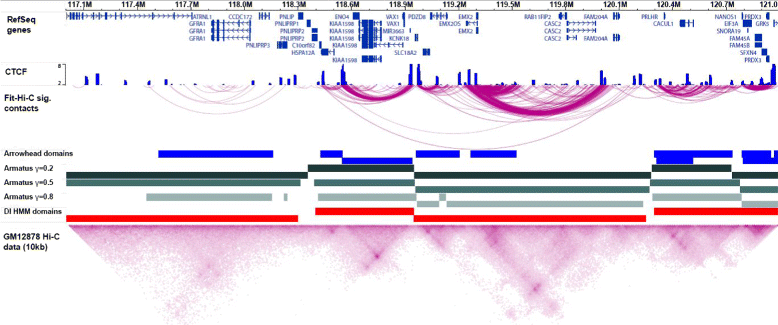
The rapidly increasing quantity of genome-wide chromosome conformation capture data presents great opportunities and challenges in the computational modeling and interpretation of the three-dimensional genome. In particular, with recent trends towards higher-resolution high-throughput chromosome conformation capture (Hi-C) data, the diversity and complexity of biological hypotheses that can be tested necessitates rigorous computational and statistical methods as well as scalable pipelines to interpret these datasets. Here we review computational tools to interpret Hi-C data, including pipelines for mapping, filtering, and normalization, and methods for confidence estimation, domain calling, visualization, and three-dimensional modeling.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

