Conserved biosynthetic pathways for phosalacine, bialaphos and newly discovered phosphonic acid natural products
- PMID: 26328935
- PMCID: PMC4731264
- DOI: 10.1038/ja.2015.77
Conserved biosynthetic pathways for phosalacine, bialaphos and newly discovered phosphonic acid natural products
Abstract
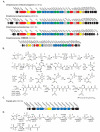
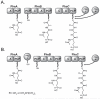
Natural products containing phosphonic or phosphinic acid functionalities often display potent biological activities with applications in medicine and agriculture. The herbicide phosphinothricin-tripeptide (PTT) was the first phosphinate natural product discovered, yet despite numerous studies, questions remain surrounding key transformations required for its biosynthesis. In particular, the enzymology required to convert phosphonoformate to carboxyphosphonoenolpyruvate and the mechanisms underlying phosphorus methylation remain poorly understood. In addition, the model for non-ribosomal peptide synthetase assembly of the intact tripeptide product has undergone numerous revisions that have yet to be experimentally tested. To further investigate the biosynthesis of this unusual natural product, we completely sequenced the PTT biosynthetic locus from Streptomyces hygroscopicus and compared it with the orthologous cluster from Streptomyces viridochromogenes. We also sequenced and analyzed the closely related phosalacine (PAL) biosynthetic locus from Kitasatospora phosalacinea. Using data drawn from the comparative analysis of the PTT and PAL pathways, we also evaluate three related recently discovered phosphonate biosynthetic loci from Streptomyces sviceus, Streptomyces sp. WM6386 and Frankia alni. Our observations address long-standing biosynthetic questions related to PTT and PAL production and suggest that additional members of this pharmacologically important class await discovery.
Figures




References
-
- De Clercq E. The Holy Trinity: the acyclic nucleoside phosphonates. Adv Pharmacol. 2013;67:293–316. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

