Gonadal transcriptomics elucidate patterns of adaptive evolution within marine rockfishes (Sebastes)
- PMID: 26329285
- PMCID: PMC4557894
- DOI: 10.1186/s12864-015-1870-0
Gonadal transcriptomics elucidate patterns of adaptive evolution within marine rockfishes (Sebastes)
Abstract
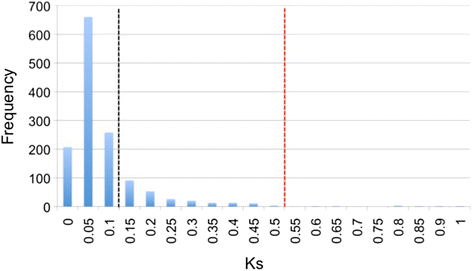
Background: The genetic mechanisms of speciation and adaptation in the marine environment are not well understood. The rockfish genus Sebastes provides a unique model system for studying adaptive evolution because of the extensive diversity found within this group, which includes morphology, ecology, and a broad range of life spans. Examples of adaptive radiations within marine ecosystems are considered an anomaly due to the absence of geographical barriers and the presence of gene flow. Using marine rockfishes, we identified signatures of natural selection from transcriptomes developed from gonadal tissue of two rockfish species (Sebastes goodei and S. saxicola). We predicted orthologous transcript pairs, and estimated their distributions of nonsynonymous (Ka) and synonymous (Ks) substitution rates.
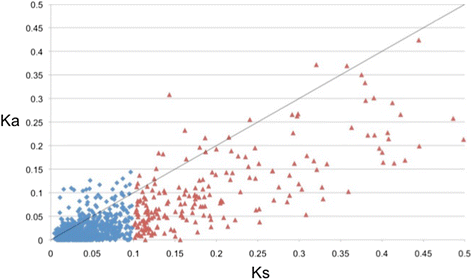
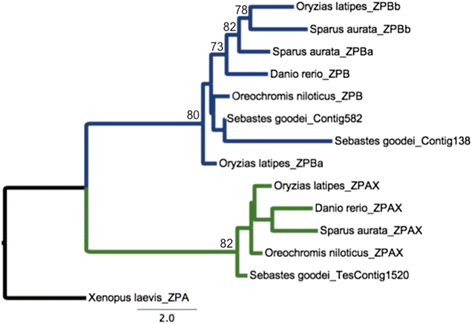
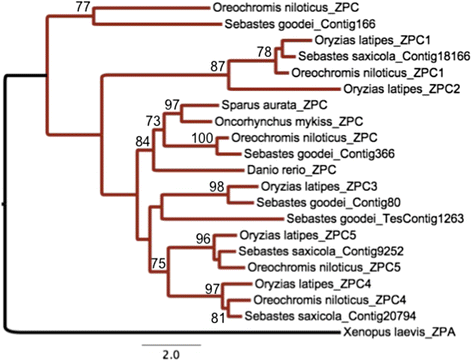
Results: We identified 144 genes out of 1079 orthologous pairs under positive selection, of which 11 are functionally annotated to reproduction based on gene ontologies (GOs). One orthologous pair of the zona pellucida gene family, which is known for its role in the selection of sperm by oocytes, out of ten was identified to be evolving under positive selection. In addition to our results in the protein coding-regions of transcripts, we found substitution rates in 3' and 5' UTRs to be significantly lower than Ks substitution rates implying negative selection in these regions.
Conclusions: We were able to identify a series of candidate genes that are useful for the assessment of the critical genes that diverged and are responsible for the radiation within this genus. Genes associated with longevity hold potential for understanding the molecular mechanisms that have contributed to the radiation within this genus.
Figures

Similar articles
-
Comparative Transcriptomics Reveals Patterns of Adaptive Evolution Associated with Depth and Age Within Marine Rockfishes (Sebastes).J Hered. 2019 May 7;110(3):340-350. doi: 10.1093/jhered/esy070. J Hered. 2019. PMID: 30602025 Free PMC article.
-
Regions of genetic divergence in depth-separated Sebastes rockfish species pairs: Depth as a potential driver of speciation.Mol Ecol. 2021 Sep;30(17):4259-4275. doi: 10.1111/mec.16046. Epub 2021 Jul 10. Mol Ecol. 2021. PMID: 34181798
-
Speciation along a depth gradient in a marine adaptive radiation.Proc Biol Sci. 2011 Feb 22;278(1705):613-8. doi: 10.1098/rspb.2010.1127. Epub 2010 Sep 1. Proc Biol Sci. 2011. PMID: 20810434 Free PMC article.
-
The geography of morphological convergence in the radiations of Pacific Sebastes rockfishes.Am Nat. 2014 Nov;184(5):E115-31. doi: 10.1086/678053. Epub 2014 Oct 6. Am Nat. 2014. PMID: 25325753
-
Probing the evolution, ecology and physiology of marine protists using transcriptomics.Nat Rev Microbiol. 2017 Jan;15(1):6-20. doi: 10.1038/nrmicro.2016.160. Epub 2016 Nov 21. Nat Rev Microbiol. 2017. PMID: 27867198 Review.
Cited by
-
Speciation genomics and the role of depth in the divergence of rockfishes (Sebastes) revealed through Pool-seq analysis of enriched sequences.Ecol Evol. 2022 Sep 23;12(9):e9341. doi: 10.1002/ece3.9341. eCollection 2022 Sep. Ecol Evol. 2022. PMID: 36188524 Free PMC article.
-
Comparative Transcriptomics Reveals Patterns of Adaptive Evolution Associated with Depth and Age Within Marine Rockfishes (Sebastes).J Hered. 2019 May 7;110(3):340-350. doi: 10.1093/jhered/esy070. J Hered. 2019. PMID: 30602025 Free PMC article.
-
Population assignment and local adaptation along an isolation-by-distance gradient in Pacific cod (Gadus macrocephalus).Evol Appl. 2018 May 23;11(8):1448-1464. doi: 10.1111/eva.12639. eCollection 2018 Sep. Evol Appl. 2018. PMID: 30151052 Free PMC article.
-
Complete Mitochondrial Genome Sequences of Five Rockfishes (Perciformes: Sebastes).Mitochondrial DNA B Resour. 2018;3(2):825-826. doi: 10.1080/23802359.2018.1495130. Epub 2018 Aug 10. Mitochondrial DNA B Resour. 2018. PMID: 31440583 Free PMC article.
-
Comparative transcriptome analysis and identification of candidate effectors in two related rust species (Gymnosporangium yamadae and Gymnosporangium asiaticum).BMC Genomics. 2017 Aug 23;18(1):651. doi: 10.1186/s12864-017-4059-x. BMC Genomics. 2017. PMID: 28830353 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

