Neuromesodermal progenitors and the making of the spinal cord
- PMID: 26329597
- PMCID: PMC4958456
- DOI: 10.1242/dev.119768
Neuromesodermal progenitors and the making of the spinal cord
Abstract
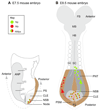
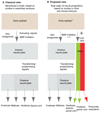
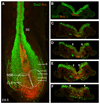
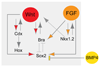
Neuromesodermal progenitors (NMps) contribute to both the elongating spinal cord and the adjacent paraxial mesoderm. It has been assumed that these cells arise as a result of patterning of the anterior neural plate. However, as the molecular mechanisms that specify NMps in vivo are uncovered, and as protocols for generating these bipotent cells from mouse and human pluripotent stem cells in vitro are established, the emerging data suggest that this view needs to be revised. Here, we review the characteristics, regulation, in vitro derivation and in vivo induction of NMps. We propose that these cells arise within primitive streak-associated epiblast via a mechanism that is separable from that which establishes neural fate in the anterior epiblast. We thus argue for the existence of two distinct routes for making central nervous system progenitors.
Keywords: Bipotent cells; FGF; Neural induction; Neuromesodermal progenitors; Spinal cord; Stem cells; Wnt.
© 2015. Published by The Company of Biologists Ltd.
Conflict of interest statement
The authors declare no competing or financial interests.
Figures


References
-
- Ang SL, Conlon RA, Jin O, Rossant J. Positive and negative signals from mesoderm regulate the expression of mouse Otx2 in ectoderm explants. Development. 1994;120:2979–2989. - PubMed
-
- Aulehla A, Wehrle C, Brand-Saberi B, Kemler R, Gossler A, Kanzler BG, Herrmann BG. Wnt3a plays a major role in the segmentation clock controlling somitogenesis. Dev Cell. 2003;4:395–406. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

