Evidence for the expression of abundant microRNAs in the locust genome
- PMID: 26329925
- PMCID: PMC4556993
- DOI: 10.1038/srep13608
Evidence for the expression of abundant microRNAs in the locust genome
Abstract
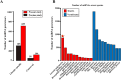
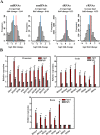
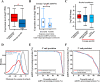
Substantial accumulation of neutral sequences accounts for genome size expansion in animal genomes. Numerous novel microRNAs (miRNAs), which evolve in a birth and death manner, are considered evolutionary neutral sequences. The migratory locust is an ideal model to determine whether large genomes contain abundant neutral miRNAs because of its large genome size. A total of 833 miRNAs were discovered, and several miRNAs were randomly chosen for validation by Northern blot and RIP-qPCR. Three additional verification methods, namely, processing-dependent methods of miRNA biogenesis using RNAi, evolutionary comparison with closely related species, and evidence supported by tissue-specific expression, were applied to provide compelling results that support the authenticity of locust miRNAs. We observed that abundant local duplication events of miRNAs, which were unique in locusts compared with those in other insects with small genome sizes, may be responsible for the substantial acquisition of miRNAs in locusts. Together, multiple evidence showed that the locust genome experienced a burst of miRNA acquisition, suggesting that genome size expansion may have considerable influences of miRNA innovation. These results provide new insight into the genomic dynamics of miRNA repertoires under genome size evolution.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

