Genome Evolution and Innovation across the Four Major Lineages of Cryptococcus gattii
- PMID: 26330512
- PMCID: PMC4556806
- DOI: 10.1128/mBio.00868-15
Genome Evolution and Innovation across the Four Major Lineages of Cryptococcus gattii
Abstract
Cryptococcus gattii is a fungal pathogen of humans, causing pulmonary infections in otherwise healthy hosts. To characterize genomic variation among the four major lineages of C. gattii (VGI, -II, -III, and -IV), we generated, annotated, and compared 16 de novo genome assemblies, including the first for the rarely isolated lineages VGIII and VGIV. By identifying syntenic regions across assemblies, we found 15 structural rearrangements, which were almost exclusive to the VGI-III-IV lineages. Using synteny to inform orthology prediction, we identified a core set of 87% of C. gattii genes present as single copies in all four lineages. Remarkably, 737 genes are variably inherited across lineages and are overrepresented for response to oxidative stress, mitochondrial import, and metal binding and transport. Specifically, VGI has an expanded set of iron-binding genes thought to be important to the virulence of Cryptococcus, while VGII has expansions in the stress-related heat shock proteins relative to the other lineages. We also characterized genes uniquely absent in each lineage, including a copper transporter absent from VGIV, which influences Cryptococcus survival during pulmonary infection and the onset of meningoencephalitis. Through inclusion of population-level data for an additional 37 isolates, we identified a new transcontinental clonal group that we name VGIIx, mitochondrial recombination between VGII and VGIII, and positive selection of multidrug transporters and the iron-sulfur protein aconitase along multiple branches of the phylogenetic tree. Our results suggest that gene expansion or contraction and positive selection have introduced substantial variation with links to mechanisms of pathogenicity across this species complex.
Importance: The genetic differences between phenotypically different pathogens provide clues to the underlying mechanisms of those traits and can lead to new drug targets and improved treatments for those diseases. In this paper, we compare 16 genomes belonging to four highly differentiated lineages of Cryptococcus gattii, which cause pulmonary infections in otherwise healthy humans and other animals. Half of these lineages have not had their genomes previously assembled and annotated. We identified 15 ancestral rearrangements in the genome and over 700 genes that are unique to one or more lineages, many of which are associated with virulence. In addition, we found evidence for recent transcontinental spread, mitochondrial genetic exchange, and positive selection in multidrug transporters. Our results suggest that gene expansion/contraction and positive selection are diversifying the mechanisms of pathogenicity across this species complex.
Copyright © 2015 Farrer et al.
Figures
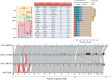

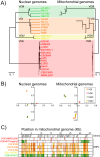
References
-
- D’Souza CA, Kronstad JW, Taylor G, Warren R, Yuen M, Hu G, Jung WH, Sham A, Kidd SE, Tangen K, Lee N, Zeilmaker T, Sawkins J, McVicker G, Shah S, Gnerre S, Griggs A, Zeng Q, Bartlett K, Li W, Wang X, Heitman J, Stajich JE, Fraser JA, Meyer W, Carter D, Schein J, Krzywinski M, Kwon-Chung KJ, Varma A, Wang J, Brunham R, Fyfe M, Ouellette BFF, Siddiqui A, Marra M, Jones S, Holt R, Birren BW, Galagan JE, Cuomo CA. 2011. Genome variation in Cryptococcus gattii, an emerging pathogen of immunocompetent hosts. mBio 2(1):e00342-10. doi: 10.1128/mBio.00342-10. - DOI - PMC - PubMed
-
- Voelz K, Ma H, Phadke S, Byrnes EJ, Zhu P, Mueller O, Farrer RA, Henk DA, Lewit Y, Hsueh Y-P, Fisher MC, Idnurm A, Heitman J, May RC. 2013. Transmission of hypervirulence traits via sexual reproduction within and between lineages of the human fungal pathogen Cryptococcus gattii. PLoS Genet 9:e1003771. doi: 10.1371/journal.pgen.1003771. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

